A Software Tool for Reagent Design to Expand Access to Single-Nucleotide Variant Detection by the Oligonucleotide Ligation Assay
- PMID: 39818318
- PMCID: PMC12179523
- DOI: 10.1016/j.jmoldx.2024.12.007
A Software Tool for Reagent Design to Expand Access to Single-Nucleotide Variant Detection by the Oligonucleotide Ligation Assay
Abstract
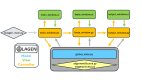
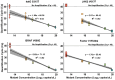
Single-nucleotide variants (SNVs) and polymorphisms are characteristic biomarkers in various biological contexts, including pathogen drug resistances and human diseases. Tools that lower the implementation barrier of molecular SNV detection methods would provide greater leverage of the expanding single-nucleotide polymorphism/SNV database. The oligonucleotide ligation assay (OLA) is a highly specific means for detection of known SNVs and is especially powerful when coupled with PCR. Yet, the OLA design process remains intensive, and criteria for success are uncertain. To assist in the design process, this study describes OLAgen, an open-source tool to automate development of OLAs and their coupled PCR assays. The software facilitates alignment of sequences surrounding SNVs and generates ligation probes while screening for dimerization potential. OLAgen successfully produced ligation probes that closely matched previously validated designs for HIV-1, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and KRAS, confirming its reliability and potential for clinical applications. The tool was used to generate new assays targeting Mycobacterium tuberculosis drug resistance and variants in the human JAK2, BRAF, and factor V genes, all of which demonstrated 100% sensitivity and specificity in controlled laboratory experiments. The OLAgen predicted assay designs detected mutant frequencies as low as 1% to 5% in wild-type backgrounds in proof-of-concept laboratory studies. OLAgen represents a significant advancement in accessible assay design, promoting the broader application of OLA technology in clinical and research settings.
Copyright © 2025 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure Statement None declared.
Figures







Similar articles
-
Surveillance for Violent Deaths - National Violent Death Reporting System, 50 States, the District of Columbia, and Puerto Rico, 2022.MMWR Surveill Summ. 2025 Jun 12;74(5):1-42. doi: 10.15585/mmwr.ss7405a1. MMWR Surveill Summ. 2025. PMID: 40493548 Free PMC article.
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
Characterization of SARS-CoV-2 intrahost genetic evolution in vaccinated and non-vaccinated patients from the Kenyan population.J Virol. 2025 Jun 17;99(6):e0048225. doi: 10.1128/jvi.00482-25. Epub 2025 May 6. J Virol. 2025. PMID: 40326760 Free PMC article.
-
Defining disease severity in atopic dermatitis and psoriasis for the application to biomarker research: an interdisciplinary perspective.Br J Dermatol. 2024 Jun 20;191(1):14-23. doi: 10.1093/bjd/ljae080. Br J Dermatol. 2024. PMID: 38419411 Free PMC article. Review.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
References
-
- Setareh M., Titov L., Surkova L. High level association of mutation in KatG315 with MDR and XDR clinical isolates of Mycobacterium tuberculosis in Belarus. Acta Microbiol Immunol Hung. 2009;56:313–325. - PubMed
-
- Babouee Flury B., Ellington M.J., Hopkins K.L., Turton J.F., Doumith M., Loy R., Staves P., Hinic V., Frei R., Woodford N. Association of novel nonsynonymous single nucleotide polymorphisms in ampD with cephalosporin resistance and phylogenetic variations in ampC, ampR, ompF, and ompC in enterobacter cloacae isolates that are highly resistant to carbapenems. Antimicrob Agents Chemother. 2016;60:2383–2390. - PMC - PubMed
-
- Clavel F., Hance A.J. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

