Proteins Involved in Endothelial Function and Inflammation Are Implicated in Cerebral Small Vessel Disease
- PMID: 39818967
- PMCID: PMC7617319
- DOI: 10.1161/STROKEAHA.124.049079
Proteins Involved in Endothelial Function and Inflammation Are Implicated in Cerebral Small Vessel Disease
Abstract
Background: Endothelial dysfunction and inflammation have been implicated in the pathophysiology of cerebral small vessel disease (SVD). However, whether they are causal, and if so which components of the pathways represent potential treatment targets, remains uncertain.
Methods: Two-sample Mendelian randomization (MR) was used to test the association between the circulating abundance of 996 proteins involved in endothelial dysfunction and inflammation and SVD. The genetic instruments predicting protein levels were obtained from the Iceland 36K (n=35 892) and the UK Biobank Proteomics (n=34 557) cohorts, both of which were longitudinal studies with follow-up from 2000 to 2023 and 2006 to 2023, respectively. SVD was represented by lacunar stroke (n=6030 cases) and 5 neuroimaging features (white matter hyperintensities [n=55 291], diffusion tensor imaging metrics: mean diffusivity [n=36 460] and fractional anisotropy [n=36 533], extensive white matter perivascular space burden [n=9324 cases], and cerebral microbleeds [n=3556 cases]). Among the proteins supported by causal evidence from the MR, cross-sectional analysis was performed to assess their associations with cognitive performance; survival analysis with Fine-Gray models was applied to examine their associations with incident all-cause dementia and stroke within the UK Biobank Proteomics cohort.
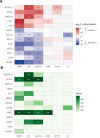
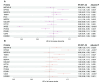
Results: MR suggested COL2A1 (collagen type II α-1 chain) was associated with lacunar stroke (odds ratio, 0.89 [95% CI, 0.86-0.91]; P=5×10-5). Moreover, 12 proteins related to endothelial function and inflammation were associated with neuroimaging features of SVD. Cross-sectional analyses showed 5 of the 13 proteins (EPHA2 [ephrin type-A receptor 2], METAP1D [methionine aminopeptidase 1D, mitochondrial], FLT4 [vascular endothelial growth factor receptor 3], COL2A1, and TIMD4 [T-cell immunoglobulin and mucin domain-containing protein 4]) were associated with cognitive performance with effects concordant with their MR findings. Survival analyses with the Fine-Gray models indicated that 5 of the 13 proteins (EPHA2, METAP1D, FLT4, APOE [apolipoprotein E], and PDE5A [cGMP-specific 3',5'-cyclic phosphodiesterase]) were associated with the risk of all-cause dementia or stroke independent of age and sex, consistent with their MR evidence.
Conclusions: Our findings suggest that endothelial-platelet activation and complement-mediated regulation of inflammation play roles in SVD and identify potential therapeutic targets and pathways.
Keywords: Mendelian randomization analysis; cerebral small vessel diseases; dementia; inflammation; stroke.
Conflict of interest statement
None.
Figures



References
-
- Duering M, Biessels GJ, Brodtmann A, Chen C, Cordonnier C, de Leeuw FE, Debette S, Frayne R, Jouvent E, Rost NS, et al. . Neuroimaging standards for research into small vessel disease—advances since 2013. Lancet Neurol. 2023;22:602–618. doi: 10.1016/S1474-4422(23)00131-X - PubMed
-
- Rajani RM, Quick S, Ruigrok SR, Graham D, Harris SE, Verhaaren BFJ, Fornage M, Seshadri S, Atanur SS, Dominiczak AF, et al. . Reversal of endothelial dysfunction reduces white matter vulnerability in cerebral small vessel disease in rats. Sci Transl Med. 2018;10:eaam9507. doi: 10.1126/scitranslmed.aam9507 - PubMed
-
- Low A, Mak E, Rowe JB, Markus HS, O’Brien JT. Inflammation and cerebral small vessel disease: a systematic review. Ageing Res Rev. 2019;53:100916. doi: 10.1016/j.arr.2019.100916 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

