Genome-wide association study and genomic prediction of root system architecture traits in Sorghum (Sorghum bicolor (L.) Moench) at the seedling stage
- PMID: 39819271
- PMCID: PMC11740658
- DOI: 10.1186/s12870-025-06077-w
Genome-wide association study and genomic prediction of root system architecture traits in Sorghum (Sorghum bicolor (L.) Moench) at the seedling stage
Abstract
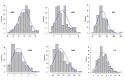
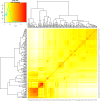
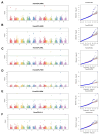
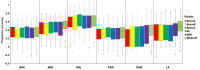
Root system architecture (RSA) plays an important role in plant adaptation to drought stress. However, the genetic basis of RSA in sorghum has not been adequately elucidated. This study aimed to investigate the genetic bases of RSA traits through genome-wide association studies (GWAS) and determine genomic prediction (GP) accuracy in sorghum landraces at the seedling stage. Phenotypic data for nodal root angle (NRA), number of nodal roots (NNR), nodal root length (NRL), fresh shoot weight (FSW), dry shoot weight (DSW), and leaf area (LA) were collected from 160 sorghum accessions grown in soil-based rhizotrons. The sorghum panel was genotyped with 5,000 single nucleotide polymorphism (SNP) markers for use in the current GWAS and GP studies. A multi-locus model, Fixed and random model Circulating Probability Unification (FarmCPU), was applied for GWAS analysis. For GP, ridge-regression best linear unbiased prediction (RR-BLUP) and five different Bayesian models were applied. A total of 17 SNP loci significantly associated with the studied traits were identified, of which nine are novel loci. Among the traits, the highest number of significant marker-trait associations (MTAs) was identified for nodal root angle on chromosomes 1, 3, 6, and 7. The SNP loci that explain the highest proportion of phenotypic variance (PVE) include sbi32853830 (PVE = 18.2%), sbi29954292 (PVE = 18.1%), sbi24668980 (PVE = 10.8%), sbi3022983 (PVE = 7%), sbi29897704 (PVE = 6.4%) and sbi29897694 (PVE = 5.3%) for the traits NNR, LA, SDW, NRA, NRL and SFW, respectively. The genomic prediction accuracy estimated for the studied traits using five Bayesian models ranged from 0.30 to 0.63 while it ranged from 0.35 to 0.60 when the RR-BLUP model was used. The observed moderate to high prediction accuracy for each trait suggests that genomic selection could be a feasible approach to sorghum RSA-targeted selection and breeding. Overall, the present study provides insights into the genetic bases of RSA and offers an opportunity to speed up breeding for drought-tolerant sorghum varieties.
Keywords: GWAS; Genomic prediction; Quantitative trait locus; Root system architecture; Sorghum.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests. Clinical trial number: Not applicable.
Figures

References
-
- McCormick RF, Truong SK, Sreedasyam A, Jenkins J, Shu S, Sims D, Kennedy M, Amirebrahimi M, Weers BD, McKinley B. The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018;93(2):338–54. - PubMed
-
- Waniska R, Rooney L, McDonough C. Sorghum: utilization. In: Encyclopedia of food grains, the world of food grains. Edited by C. Wrigley HC, K. Seetharaman, & J. Faubion, vol. 3. Oxford: Elsevier; 2016:116–123.
-
- Dykes L. Sorghum phytochemicals and their potential impact on human health. In: Sorghum. Edited by Zuo-Yu Zhao JD. New York, NY: Humana Press; 2019;1931:121–140. - PubMed
-
- FAOSTAT In. Food and Agriculture. Organization of the United Nations; 2022.
-
- Borrell A, van Oosterom E, George-Jaeggli B, Rodriguez D, Eyre J, Jordan DJ, Mace E, Singh V, Vadez V, Bell M. Sorghum. In: Crop physiology case histories for major crops. Edited by Calderini VOSaDF. London, United Kingdom: Academic Press; 2021: 196–221.
MeSH terms
LinkOut - more resources
Full Text Sources

