Empowering personalized oncology: evolution of digital support and visualization tools for molecular tumor boards
- PMID: 39819625
- PMCID: PMC11736948
- DOI: 10.1186/s12911-024-02821-8
Empowering personalized oncology: evolution of digital support and visualization tools for molecular tumor boards
Abstract
Background: Molecular tumor boards (MTBs) play a pivotal role in personalized oncology, leveraging complex data sets to tailor therapy for cancer patients. The integration of digital support and visualization tools is essential in this rapidly evolving field facing fast-growing data and changing clinical processes. This study addresses the gap in understanding the evolution of software and visualization needs within MTBs and evaluates the current state of digital support. Alignment between user requirements and software development is crucial to avoid waste of resources and maintain trust.
Methods: In two consecutive nationwide medical informatics projects in Germany, surveys and expert interviews were conducted as stage 1 (n = 14), stage 2 (n = 30), and stage 3 (n = 9). Surveys, via the SoSci Survey tool, covered participants' roles, working methods, and support needs. The second survey additionally addressed requirements for visualization solutions in molecular tumor boards. These aimed to understand diverse requirements for preparation, implementation, and documentation. Nine semi-structured expert interviews complemented quantitative findings through open discussion.
Results: Using quantitative and qualitative analyses, we show that existing digital tools may improve therapy recommendations and streamline MTB case preparation, while continuous training and system improvements are needed.
Conclusions: Our study contributes to the field by highlighting the importance of developing user-centric, customizable software solutions that can adapt to the fast-paced environment of MTBs to advance personalized oncology. In doing so, it lays the foundation for further advances in personalized medicine in oncology and points to a shift towards more efficient, technology-driven clinical decision-making processes. This research not only enriches our understanding of the integration of digital tools into MTBs, but also signals a broader shift towards technological innovation in healthcare.
Keywords: Annotation; Clinical decision support systems; Genomics; Molecular tumor board; Personalized oncology; Precision medicine; Requirements analysis; User-centered design; Visualization; cBioPortal.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The ethics committee of Friedrich-Alexander-University Erlangen-Nürnberg approved stage 1 of the study on January 24, 2022 (approval number: 22–24-ANF) and stage 2 and 3 (including small subsequent survey) on December 22, 2023 (approval number: 23–391-B). All participants provided written informed consent to participate in the study. All methods were carried out in accordance with relevant guidelines and regulations. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



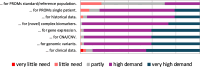
Similar articles
-
Advancing Usability and Decision Support for Molecular Tumor Boards: Insights from PM4Onco's Visual Analytics Workshop.Stud Health Technol Inform. 2025 May 15;327:512-516. doi: 10.3233/SHTI250390. Stud Health Technol Inform. 2025. PMID: 40380500
-
ESMO Precision Oncology Working Group recommendations on the structure and quality indicators for molecular tumour boards in clinical practice.Ann Oncol. 2025 Jun;36(6):614-625. doi: 10.1016/j.annonc.2025.02.009. Epub 2025 Apr 6. Ann Oncol. 2025. PMID: 40194904
-
Software-Tool Support for Collaborative, Virtual, Multi-Site Molecular Tumor Boards.SN Comput Sci. 2023;4(4):358. doi: 10.1007/s42979-023-01771-8. Epub 2023 Apr 27. SN Comput Sci. 2023. PMID: 37131499 Free PMC article.
-
Evolution of Precision Oncology, Personalized Medicine, and Molecular Tumor Boards.Surg Oncol Clin N Am. 2024 Apr;33(2):197-216. doi: 10.1016/j.soc.2023.12.004. Epub 2023 Dec 18. Surg Oncol Clin N Am. 2024. PMID: 38401905 Free PMC article. Review.
-
User Requirements for Comanaged Digital Health and Care: Review.J Med Internet Res. 2022 Jun 10;24(6):e35337. doi: 10.2196/35337. J Med Internet Res. 2022. PMID: 35687379 Free PMC article. Review.
Cited by
-
User-centered design approach to visualize PROMs for molecular tumor boards.NPJ Precis Oncol. 2025 Aug 5;9(1):273. doi: 10.1038/s41698-025-01061-x. NPJ Precis Oncol. 2025. PMID: 40764684 Free PMC article.
References
-
- Horak P, Leichsenring J, Kreutzfeldt S, Kazdal D, Teleanu V, Endris V, et al. Varianteninterpretation in der molekularen Pathologie und Onkologie: Eine Einführung. Pathologe. 2021;42(4):369–79. - PubMed
-
- Illert AL, Stenzinger A, Bitzer M, Horak P, Gaidzik VI, Möller Y, et al. The German network for personalized medicine to enhance patient care and translational research. Nat Med. 2023;29(6):1298–301. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

