Adaptive evolution of SARS-CoV-2 during a persistent infection for 521 days in an immunocompromised patient
- PMID: 39820045
- PMCID: PMC11739519
- DOI: 10.1038/s41525-025-00463-x
Adaptive evolution of SARS-CoV-2 during a persistent infection for 521 days in an immunocompromised patient
Abstract
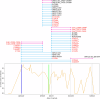
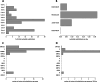
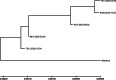
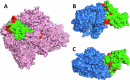
Immunocompromised patients struggle to adequately clear viral infections, offering the virus the opportunity to adapt to the immune system in the host. Here we present a case study of a patient undergoing allogeneic hematopoietic stem cell transplantation with a 521-day follow-up of a SARS-CoV-2 infection with the BF.7.21 variant. Virus samples from five time points were submitted to whole genome sequencing. Between the first detection of SARS-CoV-2 infection and its clearance, the patient's virus population acquired 34 amino acid substitutions and 8 deletions in coding regions. With 11 amino acid substitutions in the receptor binding domain of the virus' spike protein, substitutions were 15 times more abundant than expected for a random distribution in this highly functional region. Amongst them were the substitutions S:K417T, S:N440S, S:K444R, S:V445A, S:G446N, S:L452Q, S:N460K, and S:E484V at positions that are notorious for their resistance-mediating effects. The substitution patterns found indicate ongoing adaptive evolution.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Long-Term Evolution of SARS-CoV-2 in an Immunocompromised Patient with Non-Hodgkin Lymphoma.mSphere. 2021 Aug 25;6(4):e0024421. doi: 10.1128/mSphere.00244-21. Epub 2021 Jul 28. mSphere. 2021. PMID: 34319130 Free PMC article.
-
SARS-CoV-2 evolution during prolonged infection in immunocompromised patients.mBio. 2024 Mar 13;15(3):e0011024. doi: 10.1128/mbio.00110-24. Epub 2024 Feb 16. mBio. 2024. PMID: 38364100 Free PMC article.
-
SARS-CoV-2 Genome Variations in Viral Shedding of an Immunocompromised Patient with Non-Hodgkin's Lymphoma.Viruses. 2023 Jan 28;15(2):377. doi: 10.3390/v15020377. Viruses. 2023. PMID: 36851588 Free PMC article.
-
Effective virus-specific T-cell therapy for high-risk SARS-CoV-2 infections in hematopoietic stem cell transplant recipients: initial case studies and literature review.Geroscience. 2024 Feb;46(1):1083-1106. doi: 10.1007/s11357-023-00858-7. Epub 2023 Jul 6. Geroscience. 2024. PMID: 37414968 Free PMC article. Review.
-
The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern.Front Med (Lausanne). 2022 May 20;9:849217. doi: 10.3389/fmed.2022.849217. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35669924 Free PMC article. Review.
References
-
- Machkovech, H. M. et al. Persistent SARS-CoV-2 infection: significance and implications. Lancet Infect. Dis.24, e453–e462 (2024). - PubMed
-
- Vergouwe, M. & Kirby, T. Following longest known chronic SARS-CoV-2 infection of 613 days, experts highlight the risk of development of novel potentially immune-evasive SARS-CoV-2 variants due to persistent infections in immunocompromised patients. EurekAlert! https://www.eurekalert.org/news-releases/1041699 (2024).
-
- Snell, L. B. & Kirby, T. Longest known COVID-19 infection—505 days—described by UK researchers. EurekAlert! https://www.eurekalert.org/news-releases/950412 (2022).
LinkOut - more resources
Full Text Sources
Miscellaneous

