Efficacy and tolerability of neoadjuvant therapy with Talimogene laherparepvec in cutaneous basal cell carcinoma: a phase II trial (NeoBCC trial)
- PMID: 39820126
- PMCID: PMC11779647
- DOI: 10.1038/s43018-024-00879-x
Efficacy and tolerability of neoadjuvant therapy with Talimogene laherparepvec in cutaneous basal cell carcinoma: a phase II trial (NeoBCC trial)
Abstract
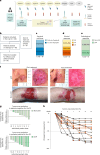
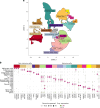
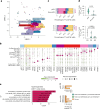
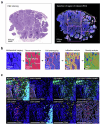
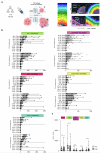
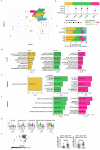
We present a single-arm, phase II, neoadjuvant trial with the oncolytic virus talimogene laherparepvec (T-VEC) in 18 patients with difficult-to-resect cutaneous basal cell carcinomas. The primary end point, defined as the proportion of patients, who after six cycles of T-VEC (13 weeks), become resectable without the need for plastic reconstructive surgery, was already achieved after stage I (9 of 18 patients; 50.0%); thus the study was discontinued for early success. The objective response rate was 55.6% and the complete pathological response rate was 33.3%. Secondary end points included safety, relapse-free survival and overall survival, time to occurrence of new basal cell carcinomas and biological read outs. Only mild adverse events occurred. The 6-month relapse-free survival and overall survival rates were 100%. In two patients a new basal cell carcinoma was diagnosed. T-VEC led to a significant increase in cytotoxic T cells (P = 0.0092), B cells (P = 0.0004) and myeloid cells (P = 0.0042) and a decrease in regulatory T cells (P = 0.0290) within the tumor microenvironment. Together, neoadjuvant T-VEC represents a viable treatment option for patients with difficult-to-resect basal cell carcinomas (EudraCT no. 2018-002165-19).
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.M.R. received speaker honoraria from Bristol-Myers Squibb, Roche, Amgen and Novartis and travel support by Sanofi, Roche and Bristol-Myers Squibb through the institution. P.T. received honoraria from Silverchair, unrestricted grants for education projects from Lilly, and honoraria for lectures from AbbVie, Lilly, FotoFinder and Novartis. J.T. reports an unrestricted grant from Lilly, consulting fees from Lilly, speaker honoraria from Lilly and Novartis, all outside the submitted work, as well as travel support from Almirall, AbbVie and Bristol-Myers Squibb through institution. C.H. reports speaker honoraria from Amgen, BMS, MSD, Novartis, Roche, participation in advisory Boards: Amgen, Astra Zeneca, BMS, Incyte, MSD, Novartis, Pierre Fabre, Roche and research funding by Amgen to the institution. The other authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

