First-in-class ultralong-target-residence-time p38α inhibitors as a mitosis-targeted therapy for colorectal cancer
- PMID: 39820127
- PMCID: PMC11864979
- DOI: 10.1038/s43018-024-00899-7
First-in-class ultralong-target-residence-time p38α inhibitors as a mitosis-targeted therapy for colorectal cancer
Abstract
Colorectal cancer (CRC) constitutes the second leading cause of cancer-related death worldwide and advanced CRCs are resistant to targeted therapies, chemotherapies and immunotherapies. p38α (Mapk14) has been suggested as a therapeutic target in CRC; however, available p38α inhibitors only allow for insufficient target inhibition. Here we describe a unique class of p38α inhibitors with ultralong target residence times (designated ULTR-p38i) that robustly inhibit p38α downstream signaling and induce distinct biological phenotypes. ULTR-p38i monotherapy triggers an uncontrolled mitotic entry by activating Cdc25 and simultaneously blocking Wee1. Consequently, CRC cells undergo mitotic catastrophe, resulting in apoptosis or senescence. ULTR-p38i exhibit high selectivity, good pharmaco-kinetic properties and no measurable toxicity with strong therapeutic effects in patient-derived CRC organoids and syngeneic CRC mouse models. Conceptually, our study suggests ultralong-target-residence-time kinase inhibitors as an alternative to covalent inhibitors, which, because of the lack of cysteine residues, cannot be generated for many kinase cancer targets.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures








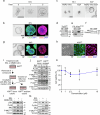

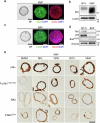


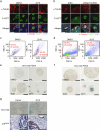
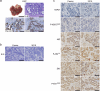
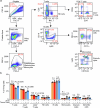
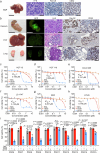
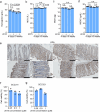
References
-
- Bedard, P. L., Hyman, D. M., Davids, M. S. & Siu, L. L. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet395, 1078–1088 (2020). - PubMed
-
- Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.74, 229–263 (2024). - PubMed
-
- Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M. & Wallace, M. B. Colorectal cancer. Lancet394, 1467–1480 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- 490941264/Deutsche Forschungsgemeinschaft (German Research Foundation)
- FOR2314/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB-TR209 - 314905040/Deutsche Forschungsgemeinschaft (German Research Foundation)
- EXC 2180 - 390900677/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB-TR240/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

