The FBXO45-GEF-H1 Axis Controls Germinal Center Formation and B-cell Lymphomagenesis
- PMID: 39820335
- PMCID: PMC11962402
- DOI: 10.1158/2159-8290.CD-24-0442
The FBXO45-GEF-H1 Axis Controls Germinal Center Formation and B-cell Lymphomagenesis
Abstract
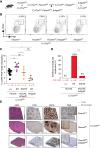
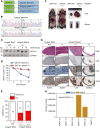
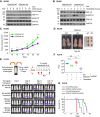
We describe the identification of a previously unrecognized ubiquitin ligase-substrate (FBXO45-GEF-H1) regulatory axis that plays an important role in germinal center formation and pathogenesis of common BCLs. These studies reveal novel insights linking dysregulated ubiquitin-mediated control to exploitable vulnerabilities and novel therapeutic strategies for these cancers.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
R. Siebert reports grants from the BMBF (Bundesministerium fur Bildung und Forschung, German Federal Ministry of Education and Research), Deutsche Krebshilfe, and DFG (Deutsche Forschungsgemeinschaft) during the conduct of the study. M. Pagano reports having been an advisor for and having financial interests in SEED Therapeutics, Triana Biomedicines, CullGen, Kymera Therapeutics, and Serinus Biosciences. M.S. Lim reports grants from the NCI during the conduct of the study and having co-founder interests in Genomenon Inc, outside the submitted work. K.S.J. Elenitoba-Johnson reports grants from the NCI and Thermo Fisher Scientific during the conduct of the study and having been an advisor for and having founder interests in Genomenon, Inc. outside the submitted work. No disclosures were reported by the other authors.
Figures




References
-
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol 2008;8:22–33. - PubMed
-
- Teras LR, DeSantis CE, Cerhan JR, Morton LM, Jemal A, Flowers CR. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin 2016;66:443–59. - PubMed
-
- Montoto S, Davies AJ, Matthews J, Calaminici M, Norton AJ, Amess J, et al. . Risk and clinical implications of transformation of follicular lymphoma to diffuse large B-cell lymphoma. J Clin Oncol 2007;25:2426–33. - PubMed
-
- Link BK, Maurer MJ, Nowakowski GS, Ansell SM, Macon WR, Syrbu SI, et al. . Rates and outcomes of follicular lymphoma transformation in the immunochemotherapy era: a report from the university of Iowa/MayoClinic Specialized Program of Research Excellence Molecular Epidemiology Resource. J Clin Oncol 2013;31:3272–8. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

