An Exosome-Based Liquid Biopsy Predicts Depth of Response and Survival Outcomes to Cetuximab and Panitumumab in Metastatic Colorectal Cancer: The EXONERATE Study
- PMID: 39820673
- PMCID: PMC11913580
- DOI: 10.1158/1078-0432.CCR-24-1934
An Exosome-Based Liquid Biopsy Predicts Depth of Response and Survival Outcomes to Cetuximab and Panitumumab in Metastatic Colorectal Cancer: The EXONERATE Study
Abstract
Purpose: The EXOsome and cell-free miRNAs of anti-EGFR ResistAnce (EXONERATE) study was an open-label, biomarker interventional study designed to develop, test, and validate a liquid biopsy predictive of progression-free survival (PFS), overall survival (OS), and objective response rate (ORR) for first-line EGFR inhibitors in metastatic colorectal cancer (mCRC).
Patients and methods: Patients with newly diagnosed RAS wild-type, chemotherapy-naïve mCRC, both right- and left-sided, were enrolled in two nationwide trials to receive cetuximab or panitumumab along with chemotherapy. The primary endpoint was 12-month PFS, which was hierarchically tested in left- and right-sided mCRCs to predict PFS, OS, and ORR.
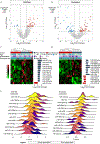
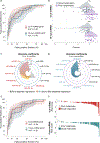
Results: Genome-wide small RNA sequencing identified 12 cell-free and 14 exosomal candidates that were differentially expressed in both plasma and tumor tissue of good versus poor responders (based on PFS <12 months). The 8 and 9 best performing candidates, respectively, were used to generate the EXONERATE assay. In left-sided mCRC, 65% were EXONERATE-high, correlating with shorter median PFS (9.5 vs. 18.5 months; P < 0.001). In the independent right-sided mCRC cohort, 80.8% were EXONERATE-high and experienced a similarly shorter median PFS (8.6 vs. 41.2 months; P = 0.0004). In the right-sided group, EXONERATE predicted PFS ≥12 months with 100% sensitivity. A linear relationship existed between EXONERATE values and response depth. Multivariate analysis revealed that EXONERATE predicts PFS and OS independently of tumor sidedness.
Conclusions: The EXONERATE assay robustly predicted PFS and OS outcomes in patients with mCRC, both right- and left-sided, before they received either panitumumab or cetuximab. It stratified PFS, OS, and ORR better than a right versus left approach.
©2025 American Association for Cancer Research.
Conflict of interest statement
Conflicts of Interest: The authors report no competing interests.
Figures



References
-
- Piawah S, Venook AP. Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 2019;125(23):4139–47. - PubMed
-
- Venook AP, Niedzwiecki D, Innocenti F, et al. Impact of primary (1°) tumor location on overall survival (OS) and progression-free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): Analysis of CALGB/SWOG 80405 (Alliance): American Society of Clinical Oncology, 2016.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

