Enhanced sensitivity and scalability with a Chip-Tip workflow enables deep single-cell proteomics
- PMID: 39820750
- PMCID: PMC11903336
- DOI: 10.1038/s41592-024-02558-2
Enhanced sensitivity and scalability with a Chip-Tip workflow enables deep single-cell proteomics
Abstract
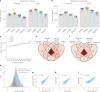
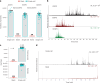
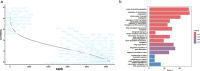
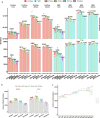
Single-cell proteomics (SCP) promises to revolutionize biomedicine by providing an unparalleled view of the proteome in individual cells. Here, we present a high-sensitivity SCP workflow named Chip-Tip, identifying >5,000 proteins in individual HeLa cells. It also facilitated direct detection of post-translational modifications in single cells, making the need for specific post-translational modification-enrichment unnecessary. Our study demonstrates the feasibility of processing up to 120 label-free SCP samples per day. An optimized tissue dissociation buffer enabled effective single-cell disaggregation of drug-treated cancer cell spheroids, refining overall SCP analysis. Analyzing nondirected human-induced pluripotent stem cell differentiation, we consistently quantified stem cell markers OCT4 and SOX2 in human-induced pluripotent stem cells and lineage markers such as GATA4 (endoderm), HAND1 (mesoderm) and MAP2 (ectoderm) in different embryoid body cells. Our workflow sets a benchmark in SCP for sensitivity and throughput, with broad applications in basic biology and biomedicine for identification of cell type-specific markers and therapeutic targets.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.B.B.-J. and N.B. are employees of Evosep Biosystems. D.H., F.I. and A.S. are employees of Cellenion SASU. H.H., M.H. and X.L. are employees of Thermo Fisher Scientific. The other authors declare no competing interests.
Figures








References
-
- Labib, M. & Kelley, S. O. Single-cell analysis targeting the proteome. Nat. Rev. Chem.4, 143–158 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

