Longitudinal host transcriptional responses to SARS-CoV-2 infection in adults with extremely high viral load
- PMID: 39820858
- PMCID: PMC11737797
- DOI: 10.1371/journal.pone.0317033
Longitudinal host transcriptional responses to SARS-CoV-2 infection in adults with extremely high viral load
Abstract
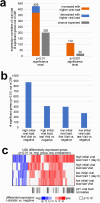
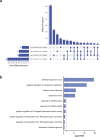
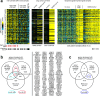
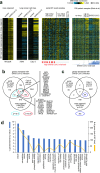
Current understanding of viral dynamics of SARS-CoV-2 and host responses driving the pathogenic mechanisms in COVID-19 is rapidly evolving. Here, we conducted a longitudinal study to investigate gene expression patterns during acute SARS-CoV-2 illness. Cases included SARS-CoV-2 infected individuals with extremely high viral loads early in their illness, individuals having low SARS-CoV-2 viral loads early in their infection, and individuals testing negative for SARS-CoV-2. We could identify widespread transcriptional host responses to SARS-CoV-2 infection that were initially most strongly manifested in patients with extremely high initial viral loads, then attenuating within the patient over time as viral loads decreased. Genes correlated with SARS-CoV-2 viral load over time were similarly differentially expressed across independent datasets of SARS-CoV-2 infected lung and upper airway cells, from both in vitro systems and patient samples. We also generated expression data on the human nose organoid model during SARS-CoV-2 infection. The human nose organoid-generated host transcriptional response captured many aspects of responses observed in the above patient samples, while suggesting the existence of distinct host responses to SARS-CoV-2 depending on the cellular context, involving both epithelial and cellular immune responses. Our findings provide a catalog of SARS-CoV-2 host response genes changing over time and magnitude of these host responses were significantly correlated to viral load.
Copyright: © 2025 Avadhanula et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Update of
-
Longitudinal host transcriptional responses to SARS-CoV-2 infection in adults with extremely high viral load.bioRxiv [Preprint]. 2023 May 25:2023.05.24.542181. doi: 10.1101/2023.05.24.542181. bioRxiv. 2023. Update in: PLoS One. 2025 Jan 16;20(1):e0317033. doi: 10.1371/journal.pone.0317033. PMID: 37292999 Free PMC article. Updated. Preprint.
-
Longitudinal host transcriptional responses to SARS-CoV-2 infection in adults with extremely high viral load.Res Sq [Preprint]. 2023 Jun 5:rs.3.rs-2978272. doi: 10.21203/rs.3.rs-2978272/v1. Res Sq. 2023. Update in: PLoS One. 2025 Jan 16;20(1):e0317033. doi: 10.1371/journal.pone.0317033. PMID: 37333115 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

