System- and sample-agnostic isotropic three-dimensional microscopy by weakly physics-informed, domain-shift-resistant axial deblurring
- PMID: 39821085
- PMCID: PMC11739688
- DOI: 10.1038/s41467-025-56078-4
System- and sample-agnostic isotropic three-dimensional microscopy by weakly physics-informed, domain-shift-resistant axial deblurring
Abstract
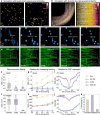
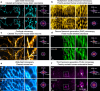
Three-dimensional subcellular imaging is essential for biomedical research, but the diffraction limit of optical microscopy compromises axial resolution, hindering accurate three-dimensional structural analysis. This challenge is particularly pronounced in label-free imaging of thick, heterogeneous tissues, where assumptions about data distribution (e.g. sparsity, label-specific distribution, and lateral-axial similarity) and system priors (e.g. independent and identically distributed noise and linear shift-invariant point-spread functions are often invalid. Here, we introduce SSAI-3D, a weakly physics-informed, domain-shift-resistant framework for robust isotropic three-dimensional imaging. SSAI-3D enables robust axial deblurring by generating a diverse, noise-resilient, sample-informed training dataset and sparsely fine-tuning a large pre-trained blind deblurring network. SSAI-3D is applied to label-free nonlinear imaging of living organoids, freshly excised human endometrium tissue, and mouse whisker pads, and further validated in publicly available ground-truth-paired experimental datasets of three-dimensional heterogeneous biological tissues with unknown blurring and noise across different microscopy systems.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources

