Brain-wide neuronal circuit connectome of human glioblastoma
- PMID: 39821165
- PMCID: PMC12347542
- DOI: 10.1038/s41586-025-08634-7
Brain-wide neuronal circuit connectome of human glioblastoma
Abstract
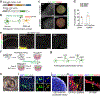
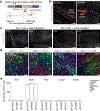
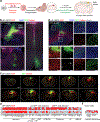
Glioblastoma (GBM) infiltrates the brain and can be synaptically innervated by neurons, which drives tumour progression1,2. Synaptic inputs onto GBM cells identified so far are largely short range and glutamatergic3,4. The extent of GBM integration into the brain-wide neuronal circuitry remains unclear. Here we applied rabies virus-mediated and herpes simplex virus-mediated trans-monosynaptic tracing5,6 to systematically investigate circuit integration of human GBM organoids transplanted into adult mice. We found that GBM cells from multiple patients rapidly integrate into diverse local and long-range neural circuits across the brain. Beyond glutamatergic inputs, we identified various neuromodulatory inputs, including synapses between basal forebrain cholinergic neurons and GBM cells. Acute acetylcholine stimulation induces long-lasting elevation of calcium oscillations and transcriptional reprogramming of GBM cells into a more motile state via the metabotropic CHRM3 receptor. CHRM3 activation promotes GBM cell motility, whereas its downregulation suppresses GBM cell motility and prolongs mouse survival. Together, these results reveal the striking capacity for human GBM cells to rapidly and robustly integrate into anatomically diverse neuronal networks of different neurotransmitter systems. Our findings further support a model in which rapid connectivity and transient activation of upstream neurons may lead to a long-lasting increase in tumour fitness.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures














Update of
-
Brain-wide neuronal circuit connectome of human glioblastoma.bioRxiv [Preprint]. 2024 Mar 4:2024.03.01.583047. doi: 10.1101/2024.03.01.583047. bioRxiv. 2024. Update in: Nature. 2025 May;641(8061):222-231. doi: 10.1038/s41586-025-08634-7. PMID: 38496540 Free PMC article. Updated. Preprint.
References
MAIN TEXT REFERENCES
-
- Venkataramani V et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573, 532–538 (2019). - PubMed
ADDITIONAL REFERENCES (METHODS)
-
- Hagemann-Jensen M et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 38, 708–714 (2020). - PubMed
-
- Kaminow B, Yunusov D & Dobin A STARsolo: accurate, fast and versatile mapping/quantification of single-cell and single-nucleus RNA-seq data. 2021.05.05.442755 Preprint at 10.1101/2021.05.05.442755 (2021). - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

