Phosphoproteomics for studying signaling pathways evoked by hormones of the renin-angiotensin system: A source of untapped potential
- PMID: 39821680
- PMCID: PMC11737475
- DOI: 10.1111/apha.14280
Phosphoproteomics for studying signaling pathways evoked by hormones of the renin-angiotensin system: A source of untapped potential
Abstract
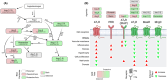
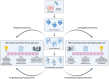
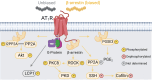
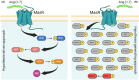
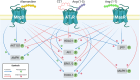
The Renin-Angiotensin System (RAS) is a complex neuroendocrine system consisting of a single precursor protein, angiotensinogen (AGT), which is processed into various peptide hormones, including the angiotensins [Ang I, Ang II, Ang III, Ang IV, Ang-(1-9), Ang-(1-7), Ang-(1-5), etc] and Alamandine-related peptides [Ang A, Alamandine, Ala-(1-5)], through intricate enzymatic pathways. Functionally, the RAS is divided into two axes with opposing effects: the classical axis, primarily consisting of Ang II acting through the AT1 receptor (AT1R), and in contrast the protective axis, which includes the receptors Mas, AT2R and MrgD and their respective ligands. A key area of RAS research is to gain a better understanding how signaling cascades elicited by these receptors lead to either "classical" or "protective" effects, as imbalances between the two axes can contribute to disease. On the other hand, therapeutic benefits can be achieved by selectively activating protective receptors and their associated signaling pathways. Traditionally, robust "hypothesis-driven" methods like Western blotting have built a solid knowledge foundation on RAS signaling. In this review, we introduce untargeted mass spectrometry-based phosphoproteomics, a "hypothesis-generating approach", to explore RAS signaling pathways. This technology enables the unbiased discovery of phosphorylation events, offering insights into previously unknown signaling mechanisms. We review the existing studies which used phosphoproteomics to study RAS signaling and discuss potential future applications of phosphoproteomics in RAS research including advantages and limitations. Ultimately, phosphoproteomics represents a so far underused tool for deepening our understanding of RAS signaling and unveiling novel therapeutic targets.
Keywords: cellular signaling; phosphoproteome; phosphoproteomics; renin‐angiotensin system.
© 2025 The Author(s). Acta Physiologica published by John Wiley & Sons Ltd on behalf of Scandinavian Physiological Society.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The vasoprotective axes of the renin-angiotensin system: Physiological relevance and therapeutic implications in cardiovascular, hypertensive and kidney diseases.Pharmacol Res. 2017 Nov;125(Pt A):21-38. doi: 10.1016/j.phrs.2017.06.005. Epub 2017 Jun 12. Pharmacol Res. 2017. PMID: 28619367 Free PMC article. Review.
-
A century old renin-angiotensin system still grows with endless possibilities: AT1 receptor signaling cascades in cardiovascular physiopathology.Cell Signal. 2014 Oct;26(10):2147-60. doi: 10.1016/j.cellsig.2014.06.011. Epub 2014 Jul 5. Cell Signal. 2014. PMID: 25007996 Review.
-
ACE2 and vasoactive peptides: novel players in cardiovascular/renal remodeling and hypertension.Ther Adv Cardiovasc Dis. 2015 Aug;9(4):217-37. doi: 10.1177/1753944715597623. Epub 2015 Aug 13. Ther Adv Cardiovasc Dis. 2015. PMID: 26275770 Review.
-
The Renin-Angiotensin System and the Neurodegenerative Diseases: A Brief Review.Protein Pept Lett. 2017 Nov 17;24(9):841-853. doi: 10.2174/0929866524666170822120258. Protein Pept Lett. 2017. PMID: 28828974 Review.
-
Alamandine: a new member of the angiotensin family.Curr Opin Nephrol Hypertens. 2014 Mar;23(2):130-4. doi: 10.1097/01.mnh.0000441052.44406.92. Curr Opin Nephrol Hypertens. 2014. PMID: 24389733 Review.
Cited by
-
G-Protein-Coupled Receptors in Chronic Kidney Disease Induced by Hypertension and Diabetes.Cells. 2025 May 16;14(10):729. doi: 10.3390/cells14100729. Cells. 2025. PMID: 40422232 Free PMC article. Review.
References
-
- Dzau VJ. Theodore cooper lecture: tissue angiotensin and pathobiology of vascular disease: a unifying hypothesis. Hypertension. 2001;37(4):1047‐1052. - PubMed
-
- Tigerstedt R, Bergman PQ. Niere und Kreislauf. Skandinavisches Archiv Für Physiologie. 1898;8(1):223‐271.
Publication types
MeSH terms
Substances
Grants and funding
- 6239/Novo Nordisk Fonden
- 0058592/Novo Nordisk Fonden
- 4004-00485B/Danmarks Frie Forskningsfond
- 0134-00297B/Danmarks Frie Forskningsfond
- BPD-00133-22/Fundação de Amparo à Pesquisa do Estado de Minas Gerais
- 88881.700905/2022-01/Coordenação de Aperfeiçoamento de Pessoal de Nível Superior
- 88887.916694/2023-00/Coordenação de Aperfeiçoamento de Pessoal de Nível Superior
- 406936/2023-4/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- 309965/2022-5/Conselho Nacional de Desenvolvimento Científico e Tecnológico
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

