FAM136A depletion induces mitochondrial stress and reduces mitochondrial membrane potential and ATP production
- PMID: 39821719
- PMCID: PMC12051016
- DOI: 10.1002/2211-5463.13967
FAM136A depletion induces mitochondrial stress and reduces mitochondrial membrane potential and ATP production
Abstract
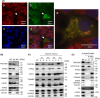
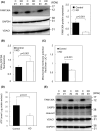
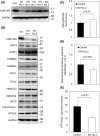
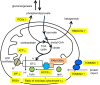
FAM136A deficiency has been associated with Ménière's disease. However, the underlying mechanism of action of this protein remains unclear. We hypothesized that FAM136A functions in maintaining mitochondria, even in HepG2 cells. To better characterize FAM136A function, we analyzed the cellular response caused by its depletion. FAM136A depletion induced reactive oxygen species (ROS) and reduced both mitochondrial membrane potential and ATP production. However, cleaved caspase-9 levels did not increase significantly. We next investigated why the depletion of FAM136A reduced the mitochondrial membrane potential and ATP production but did not lead to apoptosis. Depletion of FAM136A induced the mitochondrial unfolded protein response (UPRmt) and the expression levels of gluconeogenic phosphoenolpyruvate carboxykinases (PCK1 and PCK2) and ketogenic 3-hydroxy-3-methylglutaryl-CoA synthases (HMGCS1 and HMGCS2) were upregulated. Furthermore, depletion of FAM136A reduced accumulation of holocytochrome c synthase (HCCS), a FAM136A interacting enzyme that combines heme to apocytochrome c to produce holocytochrome c. Notably, the amount of heme in cytochrome c did not change significantly with FAM136A depletion, although the amount of total cytochrome c protein increased significantly. This observation suggests that greater amounts of cytochrome c remain unbound to heme in FAM136A-depleted cells.
Keywords: ATP; FAM136A; holocytochrome c synthetase; mitochondrial membrane potential; mitochondrial stress.
© 2025 The Author(s). FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare that they have no conflicts of interest regarding the content of this article.
Figures




Similar articles
-
ABCB10 depletion reduces unfolded protein response in mitochondria.Biochem Biophys Res Commun. 2017 Apr 29;486(2):465-469. doi: 10.1016/j.bbrc.2017.03.063. Epub 2017 Mar 16. Biochem Biophys Res Commun. 2017. PMID: 28315685
-
Mitochondria-dependent reactive oxygen species-mediated programmed cell death induced by 3,3'-diindolylmethane through inhibition of F0F1-ATP synthase in unicellular protozoan parasite Leishmania donovani.Mol Pharmacol. 2008 Nov;74(5):1292-307. doi: 10.1124/mol.108.050161. Epub 2008 Aug 14. Mol Pharmacol. 2008. PMID: 18703668
-
Brusatol Protects HepG2 Cells against Oxygen-Glucose Deprivation-Induced Injury via Inhibiting Mitochondrial Reactive Oxygen Species-Induced Oxidative Stress.Pharmacology. 2020;105(7-8):416-423. doi: 10.1159/000504482. Epub 2019 Dec 11. Pharmacology. 2020. PMID: 31825932
-
Revisiting Kadenbach: Electron flux rate through cytochrome c-oxidase determines the ATP-inhibitory effect and subsequent production of ROS.Bioessays. 2016 Jun;38(6):556-67. doi: 10.1002/bies.201600043. Epub 2016 May 12. Bioessays. 2016. PMID: 27171124 Free PMC article. Review.
-
Stress-mediated generation of deleterious ROS in healthy individuals - role of cytochrome c oxidase.J Mol Med (Berl). 2020 May;98(5):651-657. doi: 10.1007/s00109-020-01905-y. Epub 2020 Apr 20. J Mol Med (Berl). 2020. PMID: 32313986 Free PMC article. Review.
References
-
- Busch JD, Fielden LF, Pfanner N and Wiedemann N (2013) Mitochondrial protein transport: versatility of translocases and mechanisms. Mol Cell 83, 890–910. - PubMed
-
- Pfanner N and Wiedemann N (2002) Mitochondrial protein import: two membranes, three translocases. Curr Opin Cell Biol 14, 400–411. - PubMed
-
- Yamano K, Yatsukawa Y, Esaki M, Hobbs AEA, Jensen RE and Endo T (2008) Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J Biol Chem 283, 3799–3807. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

