Mutation Analysis of TMB-High Colorectal Cancer: Insights Into Molecular Pathways and Clinical Implications
- PMID: 39822019
- PMCID: PMC11967252
- DOI: 10.1111/cas.16455
Mutation Analysis of TMB-High Colorectal Cancer: Insights Into Molecular Pathways and Clinical Implications
Abstract
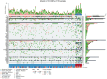
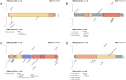
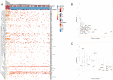
Colorectal cancer (CRC) is well characterized in terms of genetic mutations and the mechanisms by which they contribute to carcinogenesis. Mutations in APC, TP53, and KRAS are common in CRC, indicating key roles for these genes in tumor development and progression. However, for certain tumors with low frequencies of these mutations that are defined by tumor location and molecular phenotypes, a carcinogenic mechanism dependent on BRAF mutations has been proposed. We here analyzed targeted sequence data linked to clinical information for CRC, focusing on tumors with a high tumor mutation burden (TMB) in order to identify the characteristics of associated mutations, their relations to clinical features, and the mechanisms of carcinogenesis in tumors lacking the major driver oncogenes. Analysis of overall mutation frequencies confirmed that APC, TP53, and KRAS mutations were the most prevalent in our cohort. Compared with other tumors, TMB-high tumors were more frequent on the right side of the colon, had lower KRAS and higher BRAF mutation frequencies as well as a higher microsatellite instability (MSI) score, and showed a greater contribution of a mutational signature associated with MSI. Ranking of variant allele frequencies to identify genes that play a role early in carcinogenesis suggested that mutations in genes related to the DNA damage response (such as ATM and POLE) and to MSI (such as MSH2 and MSH6) may precede BRAF mutations associated with activation of the serrated pathway in TMB-high tumors. Our results thus indicate that TMB-high tumors suggest that mutations of genes related to mismatch repair and the DNA damage response may contribute to activation of the serrated pathway in CRC.
Keywords: CpG island methylator phenotype (CIMP); colorectal cancer; molecular phenotype; precision medicine; tumor mutation burden (TMB).
© 2025 The Author(s). Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
Nishihara, Hiroshi and Saya, Hideyuki is an editorial board member of Cancer Science. Other authors do not have a conflicts of interest.
Figures

References
-
- Jauhri M., Bhatnagar A., Gupta S., et al., “Prevalence and Coexistence of KRAS, BRAF, PIK3CA, NRAS, TP53, and APC Mutations in Indian Colorectal Cancer Patients: Next‐Generation Sequencing–Based Cohort Study,” Tumor Biology 39, no. 2 (2017): 1010428317692265, 10.1177/1010428317692265. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

