Pellino 3 E3 ligase promotes starvation-induced autophagy to prevent hepatic steatosis
- PMID: 39823344
- PMCID: PMC11740972
- DOI: 10.1126/sciadv.adr2450
Pellino 3 E3 ligase promotes starvation-induced autophagy to prevent hepatic steatosis
Abstract
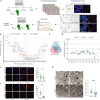
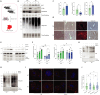
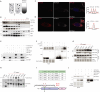
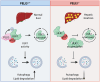
Nutrient deprivation is a major trigger of autophagy, a conserved quality control and recycling process essential for cellular and tissue homeostasis. In a high-content image-based screen of the human ubiquitome, we here identify the E3 ligase Pellino 3 (PELI3) as a crucial regulator of starvation-induced autophagy. Mechanistically, PELI3 localizes to autophagic membranes, where it interacts with the ATG8 proteins through an LC3-interacting region (LIR). This facilitates PELI3-mediated ubiquitination of ULK1, driving ULK1's subsequent proteasomal degradation. PELI3 depletion leads to an aberrant accumulation and mislocalization of ULK1 and disrupts the early steps of autophagosome formation. Genetic deletion of Peli3 in mice impairs fasting-induced autophagy in the liver and enhances starvation-induced hepatic steatosis by reducing autophagy-mediated clearance of lipid droplets. Notably, PELI3 expression is decreased in the livers of patients with metabolic dysfunction-associated steatotic liver disease (MASLD), suggesting its role in hepatic steatosis development in humans. The findings suggest that PELI3-mediated control of autophagy plays a protective role in liver health.
Figures



References
-
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N., The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004). - PubMed
-
- Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.-L., Monitoring autophagy by electron microscopy in mammalian cells. Methods Enzymol. 452, 143–164 (2009). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

