Discovery of a DNA methylation profile in individuals with Sifrim-Hitz-Weiss syndrome
- PMID: 39824190
- PMCID: PMC11866970
- DOI: 10.1016/j.ajhg.2024.12.020
Discovery of a DNA methylation profile in individuals with Sifrim-Hitz-Weiss syndrome
Abstract
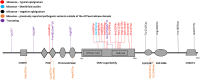
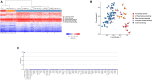
Pathogenic heterozygous variants in CHD4 cause Sifrim-Hitz-Weiss syndrome, a neurodevelopmental disorder associated with brain anomalies, heart defects, macrocephaly, hypogonadism, and additional features with variable expressivity. Most individuals have non-recurrent missense variants, complicating variant interpretation. A few were reported with truncating variants, and their role in disease is unclear. DNA methylation episignatures have emerged as highly accurate diagnostic biomarkers in a growing number of rare diseases. We aimed to study evidence for the existence of a CHD4-related DNA methylation episignature. We collected blood DNA samples and/or clinical information from 39 individuals with CHD4 variants, including missense and truncating variants. Genomic DNA methylation analysis was performed on 28 samples. We identified a sensitive and specific DNA methylation episignature in samples with pathogenic missense variants within the ATPase/helicase domain. The same episignature was observed in a family with variable expressivity, a de novo variant near the PHD domain, variants of uncertain significance within the ATPase/helicase domain, and a sample with compound heterozygous variants. DNA methylation data revealed higher percentages of shared probes with BAFopathies, CHD8, and the terminal ADNP variants encoding a protein known to form the ChAHP complex with CHD4. Truncating variants, as well as a sample with a recurrent pathogenic missense variant, exhibited DNA methylation profiles distinct from the ATPase/helicase domain episignature. These DNA methylation differences, together with the distinct clinical features observed in those individuals, provide preliminary evidence for clinical and molecular sub-types in the CHD4-related disorder.
Keywords: ADNP; CHD4; autism; chromatin remodeler; compound heterozygous; methylation; neurodevelopmental; truncating; variable expressivity.
Copyright © 2024 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.S. is a shareholder in EpiSign, Inc., a company involved in commercial uses of EpiSign technology. I.M.W. is an employee of GeneDx, LLC.
Figures




References
-
- Aref-Eshghi E., Kerkhof J., Pedro V.P., Barat-Houari M., Ruiz-Pallares N., Andrau J.C., Lacombe D., Van-Gils J., Fergelot P., et al. Groupe DI France Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020;106:356–370. doi: 10.1016/j.ajhg.2020.01.019. - DOI - PMC - PubMed
-
- Aref-Eshghi E., Rodenhiser D.I., Schenkel L.C., Lin H., Skinner C., Ainsworth P., Paré G., Hood R.L., Bulman D.E., Kernohan K.D., et al. Genomic DNA Methylation Signatures Enable Concurrent Diagnosis and Clinical Genetic Variant Classification in Neurodevelopmental Syndromes. Am. J. Hum. Genet. 2018;102:156–174. doi: 10.1016/j.ajhg.2017.12.008. - DOI - PMC - PubMed
-
- van Jaarsveld R.H., Reilly J., Cornips M.C., Hadders M.A., Agolini E., Ahimaz P., Anyane-Yeboa K., Bellanger S.A., van Binsbergen E., van den Boogaard M.J., et al. Delineation of a KDM2B-related neurodevelopmental disorder and its associated DNA methylation signature. Genet. Med. 2023;25:49–62. doi: 10.1016/j.gim.2022.09.006. - DOI - PMC - PubMed
-
- Choufani S., Gibson W.T., Turinsky A.L., Chung B.H.Y., Wang T., Garg K., Vitriolo A., Cohen A.S.A., Cyrus S., Goodman S., et al. DNA Methylation Signature for EZH2 Functionally Classifies Sequence Variants in Three PRC2 Complex Genes. Am. J. Hum. Genet. 2020;106:596–610. doi: 10.1016/j.ajhg.2020.03.008. - DOI - PMC - PubMed
-
- Awamleh Z., Choufani S., Cytrynbaum C., Alkuraya F.S., Scherer S., Fernandes S., Rosas C., Louro P., Dias P., Neves M.T., et al. ANKRD11 pathogenic variants and 16q24.3 microdeletions share an altered DNA methylation signature in patients with KBG syndrome. Hum. Mol. Genet. 2023;32:1429–1438. doi: 10.1093/hmg/ddac289. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

