Improving polyketide biosynthesis by rescuing the translation of truncated mRNAs into functional polyketide synthase subunits
- PMID: 39824802
- PMCID: PMC11742023
- DOI: 10.1038/s41467-025-55973-0
Improving polyketide biosynthesis by rescuing the translation of truncated mRNAs into functional polyketide synthase subunits
Abstract
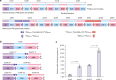
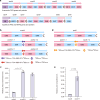
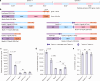
Modular polyketide synthases (mPKSs) are multidomain enzymes in bacteria that synthesize a variety of pharmaceutically important compounds. mPKS genes are usually longer than 10 kb and organized in operons. To understand the transcriptional and translational characteristics of these large genes, here we split the 13-kb busA gene, encoding a 456-kDa three-module PKS for butenyl-spinosyn biosynthesis, into three smaller separately translated genes encoding one PKS module in an operon. Expression of the native and split busA genes in Streptomyces albus reveals that the majority ( >93%) of PKS mRNAs are truncated, resulting in a greater abundance of and a higher synthesis rate for the proteins encoded by genes closer to the operon promoter. Splitting the large busA gene rescues translation of truncated mRNAs into functional PKS subunits, and increases the biosynthetic efficiency of butenyl-spinosyn PKS by 13-fold. The truncated mRNA translation rescue strategy will facilitate engineering of multi-domain proteins to enhance their functions.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: H.W., Y.L. and J.L. are inventors of the pending patent application submitted by Shandong University that covers the PKS module splitting method described in this study (CN2024118692176). The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

