Interferon-α promotes HLA-B-restricted presentation of conventional and alternative antigens in human pancreatic β-cells
- PMID: 39824805
- PMCID: PMC11748642
- DOI: 10.1038/s41467-025-55908-9
Interferon-α promotes HLA-B-restricted presentation of conventional and alternative antigens in human pancreatic β-cells
Abstract
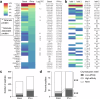
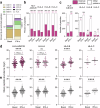
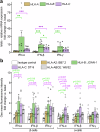
Interferon (IFN)-α is the earliest cytokine signature observed in individuals at risk for type 1 diabetes (T1D), but the effect of IFN-α on the antigen repertoire of HLA Class I (HLA-I) in pancreatic β-cells is unknown. Here we characterize the HLA-I antigen presentation in resting and IFN-α-exposed β-cells and find that IFN-α increases HLA-I expression and expands peptide repertoire to those derived from alternative mRNA splicing, protein cis-splicing and post-translational modifications. While the resting β-cell immunopeptidome is dominated by HLA-A-restricted peptides, IFN-α largely favors HLA-B and only marginally upregulates HLA-A, translating into increased HLA-B-restricted peptide presentation and activation of HLA-B-restricted CD8+ T cells. Lastly, islets of patients with T1D show preferential HLA-B hyper-expression when compared with non-diabetic donors, and islet-infiltrating CD8+ T cells reactive to HLA-B-restricted granule peptides are found in T1D donors. Thus, the inflammatory milieu of insulitis may skew the autoimmune response toward alternative epitopes presented by HLA-B, hence recruiting T cells with a distinct repertoire that may be relevant to T1D pathogenesis.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Marroqui, L. et al. Interferon-alpha mediates human beta cell HLA class I overexpression, endoplasmic reticulum stress and apoptosis, three hallmarks of early human type 1 diabetes. Diabetologia60, 656–667 (2017). - PubMed
MeSH terms
Substances
Grants and funding
- R01 DK133881/DK/NIDDK NIH HHS/United States
- EFSD/JDRF/Lilly European Programme in Type 1 Diabetes Research 2019/European Foundation for the Study of Diabetes (EFSD)
- 1901-03689, 2018PG-T1D053/Leona M. and Harry B. Helmsley Charitable Trust (Helmsley Charitable Trust)
- UC4 DK098085/DK/NIDDK NIH HHS/United States
- UC4 DK116284/DK/NIDDK NIH HHS/United States
- P30 DK116073/DK/NIDDK NIH HHS/United States
- R01 DK126444/DK/NIDDK NIH HHS/United States
- U01 DK127786/DK/NIDDK NIH HHS/United States
- EFSD/Lilly Young Investigator Research Award Programme 2023/European Foundation for the Study of Diabetes (EFSD)
- R01 DK099317/DK/NIDDK NIH HHS/United States
- EQU20193007831/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- 3-SRA-2022-1201-S-B, 5-SRA-2018-557-Q-R, Postdoctoral Fellowship 3-PDF-2020-942-A-N/JDRF/United States
- U01 DK104218/DK/NIDDK NIH HHS/United States
- ANR-19-CE15-0014-01/Agence Nationale de la Recherche (French National Research Agency)
LinkOut - more resources
Full Text Sources
Research Materials

