Integrative genetics and multiomics analysis reveal mechanisms and therapeutic targets in vitiligo highlighting JAK STAT pathway regulation of CTSS
- PMID: 39824912
- PMCID: PMC11742684
- DOI: 10.1038/s41598-025-86134-4
Integrative genetics and multiomics analysis reveal mechanisms and therapeutic targets in vitiligo highlighting JAK STAT pathway regulation of CTSS
Abstract
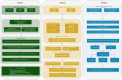
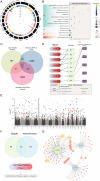
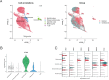
Vitiligo is a complex autoimmune disease characterized by the loss of melanocytes, leading to skin depigmentation. Despite advances in understanding its genetic and molecular basis, the precise mechanisms driving vitiligo remain elusive. Integrating multiple layers of omics data can provide a comprehensive view of disease pathogenesis and identify potential therapeutic targets. The study aims to delineate the genetic and molecular mechanisms of vitiligo pathogenesis using an integrative multiomics strategy. We focus on exploring the regulatory influence of the JAK/STAT pathway on Cathepsin S, a potential therapeutic target in vitiligo. Our GWAS-meta analysis pinpointed five druggable genes: ERBB3, RHOH, CDK10, MC1R, and NDUFAF3, and underwent drug target exploration and molecular docking. SMR analysis linked CTSS, CTSH, STX8, KIR2DL3, and GRHPR to vitiligo through pQTL and eQTL associations. Microarray and single-cell RNA-seq data showed differential expression of CTSS and STAT1/3 in vitiligo patients' blood and skin lesions. Our study offers novel perspectives on vitiligo's genetic and molecular basis, highlighting the JAK/STAT pathway's role in regulating CTSS for antigen processing in melanocytes. Further research is needed to confirm these results and assess the therapeutic potential of CTSS and related genes.
Keywords: Bioinformatics; CTSS; GWAS; Multiomics; Therapeutic targets; Vitiligo.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics statement: The data used in this study were derived from previously published research, which obtained ethical approval from their respective committees. Therefore, no additional ethical permission was required for our study. Consent for publication: In the manuscript, consent was obtained from each individual for any form of personal data included.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

