Protocol for identifying Mycobacterium tuberculosis infection status through airway microbiome profiling
- PMID: 39826114
- PMCID: PMC11787526
- DOI: 10.1016/j.xpro.2024.103574
Protocol for identifying Mycobacterium tuberculosis infection status through airway microbiome profiling
Abstract
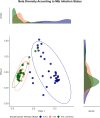
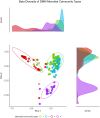
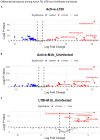
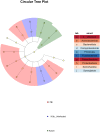
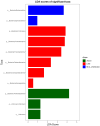
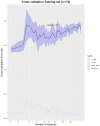
This protocol describes the steps to determine an airway microbiome signature for identifying Mycobacterium tuberculosis infection status. We outline procedures for processing microbiome data, calculating diversity measures, and fitting Dirichlet multinomial mixture models. Additionally, we provide steps for analyzing taxonomic relative and differential abundances, as well as identifying potential biomarkers associated with infection status. For complete details on the use and execution of this protocol, please refer to Kayongo et al.1.
Keywords: bioinformatics; immunology; microbiology.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures








References
-
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

