Structural development of the HIV-1 apex-directed PGT145-PGDM1400 antibody lineage
- PMID: 39826122
- PMCID: PMC11883830
- DOI: 10.1016/j.celrep.2024.115223
Structural development of the HIV-1 apex-directed PGT145-PGDM1400 antibody lineage
Abstract
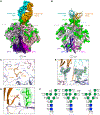
Broadly neutralizing antibodies (bNAbs) targeting the apex of the HIV-1-envelope (Env) trimer comprise the most potent category of HIV-1 bNAbs and have emerged as promising therapeutics. Here, we investigate the development of the HIV-1 apex-directed PGT145-PGDM1400 antibody lineage and report cryo-EM structures at 3.4 Å resolution of PGDM1400 and of an improved PGT145 variant (PGT145-R100aS), each bound to the BG505 Env trimer. Cross-species-based engineering improves PGT145 IC80 breadth to near that of PGDM1400. Despite similar breadth and potency, the two antibodies differ in their residue-level interactions with important apex features, including N160 glycans and apex cavity, with residue 100i of PGT145 (sulfated tyrosine) penetrating ∼7 Å farther than residue 100i of PGDM1400 (aspartic acid). While apex-directed bNAbs from other donors use maturation pathways that often converge on analogous residue-level recognition, our results demonstrate that divergent residue-level recognition can occur within the same lineage, thereby enabling improved coverage of escape variants.
Keywords: CAP256-VRC26.25; CP: Immunology; HIV-1 envelope trimer; PCT64; PG9; RHA1.V2.01; V2-apex antibody; antibody evolution; bNAb; broadly neutralizing antibody; tyrosine sulfation.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, and Burton DR (2010). A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6, e1001028. 10.1371/journal.ppat.1001028. - DOI - PMC - PubMed
-
- Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, Ramos A, Bian CB, Simek M, Allen S, et al. (2016). Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort. PLoS Pathog. 12, e1005369. 10.1371/journal.ppat.1005369. - DOI - PMC - PubMed
-
- Willis JR, Berndsen ZT, Ma KM, Steichen JM, Schiffner T, Landais E, Liguori A, Kalyuzhniy O, Allen JD, Baboo S, et al. (2022). Human immunoglobulin repertoire analysis guides design of vaccine priming immunogens targeting HIV V2-apex broadly neutralizing antibody precursors. Immunity 55, 2149–2167. 10.1016/j.immuni.2022.09.001. - DOI - PMC - PubMed
-
- Voss JE, Andrabi R, McCoy LE, de Val N, Fuller RP, Messmer T, Su CY, Sok D, Khan SN, Garces F, et al. (2017). Elicitation of Neutralizing Antibodies Targeting the V2 Apex of the HIV Envelope Trimer in a Wild-Type Animal Model. Cell Rep. 21, 222–235. 10.1016/j.celrep.2017.09.024. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

