Fiber- and acetate-mediated modulation of MHC-II expression on intestinal epithelium protects from Clostridioides difficile infection
- PMID: 39826540
- PMCID: PMC11974464
- DOI: 10.1016/j.chom.2024.12.017
Fiber- and acetate-mediated modulation of MHC-II expression on intestinal epithelium protects from Clostridioides difficile infection
Abstract
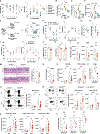
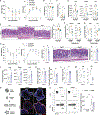
Here, we explore the relationship between dietary fibers, colonic epithelium major histocompatibility complex class II (MHC-II) expression, and immune cell interactions in regulating susceptibility to Clostridioides difficile infection (CDI). We find that a low-fiber diet increases MHC-II expression in the colonic epithelium, which, in turn, worsens CDI by promoting the development of pathogenic CD4+ intraepithelial lymphocytes (IELs). The influence of dietary fibers on MHC-II expression is mediated by its metabolic product, acetate, and its receptor, free fatty acid receptor 2 (FFAR2). While acetate activation of FFAR2 on epithelial cells helps resist CDI, it does not directly regulate MHC-II expression. Instead, MHC-II is regulated by FFAR2 in type 3 innate lymphoid cells (ILC3s). Acetate enhances interleukin-22 (IL-22) production by ILC3s, which then suppresses MHC-II expression on the colonic epithelium. In conclusion, a low-fiber diet reduces acetate-induced IL-22 production by ILC3s, leading to increased MHC-II on the colonic epithelium. This change affects recovery from CDI by expanding the population of pathogenic CD4+ IELs.
Keywords: Clostridioides difficile; MHC-II; diet; fibers; group 3 innate lymphoid cells; gut microbiota; interleukin-22; intestinal epithelial cells; intraepithelial lymphocytes; short-chain fatty acids.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Lawley TD, Clare S, Deakin LJ, Goulding D, Yen JL, Raisen C, Brandt C, Lovell J, Cooke F, Clark TG, et al. (2010). Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl. Environ. Microbiol. 76, 6895–6900. 10.1128/AEM.00718-10. - DOI - PMC - PubMed
-
- Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, Huffnagle GB, Z Li J, and Young VB (2014). Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5, 3114. 10.1038/ncomms4114. - DOI - PMC - PubMed
-
- Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, et al. (2015). Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the Host Response during Clostridium difficile Infections. mBio 6, e00551. 10.1128/mBio.00551-15. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

