Metagenomics: a new frontier for routine pathology testing of gastrointestinal pathogens
- PMID: 39827146
- PMCID: PMC11742996
- DOI: 10.1186/s13099-024-00673-1
Metagenomics: a new frontier for routine pathology testing of gastrointestinal pathogens
Abstract
Background: Accurate and comprehensive identification of enteropathogens, causing infectious gastroenteritis, is essential for optimal patient treatment and effective isolation processes in health care systems. Traditional diagnostic techniques are well established and optimised in low-cost formats. However, thorough testing for a wider range of causal agents is time consuming and remains limited to a subset of pathogenic organisms. Metagenomic next-generation sequencing (mNGS) allows the identification of all pathogens in a sample in a single test, without a reliance on culture or introduction of target selection bias. This study aims to determine the ability to routinely apply mNGS testing, in comparison to traditional culture or polymerase chain reaction (PCR) based tests, for the identification of causal pathogens for gastrointestinal infections.
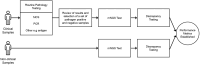
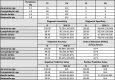
Results: The performance of mNGS, PCR and microscopy, culture and sensitivity (MCS) assays was established using 2,619 prospectively collected faecal samples from patients with symptomology indicative of infectious gastroenteritiss. Commonly experienced pathogens including Aeromonas spp, Campylobacter spp, Salmonella spp and Giardia spp, in single and co-infected patients, were used to establish test outcomes. When testing for these organisms, using the combined result from either or both PCR and MCS testing as the comparator, the mNGS assay had clinically acceptable sensitivity (89.2-100%). Further, the mNGS assay detected 14 additional enteropathogens, that were either not detected or not tested, by initial PCR/MCS testing.
Conclusions: The advantage of mNGS compared to other syndromic testing systems is the broad range of detectable targets and the ability to interrogate samples without clinician informed or assay specific bias. With the development of newer sequencing assays, it is now feasible to test for a wide range of target organisms in a sample using a single mNGS test. Overall, the mNGS based approach enabled pathogen detection that was comparable to conventional diagnostics and was shown to have the potential to be extended for the detection of many pathogens and genes of clinical interest. In conclusion, the mNGS assay offers an easy, sample to answer workflow with rapid detection of enteropathogens and has the potential to improve diagnosis, therapy and infection control precautions.
Keywords: Enteropathogens; Faecal; Gastrointestinal; Infection; Metagenomics; Pathogen.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All participants provided informed consent within the approved research project by the national ethics committee of Bellberry Limited (Project No.: 2018–05-400-A-2) and local ethics committee, Mater Research Clinical Governance Office (Project 46934 Ref AM/MML/46934 v4). Consent for publication: Not applicable. Competing interests: All authors are shareholders and/or receive a salary or reimbursement from Microba Pty Ltd. It is noted that the reported outcomes in the paper are directly from validation of a metagenomics assay for the detection of infectious gastrointestinal organisms, which has been independently reviewed and assessed by an external certification body to ISO15189, as being true and ubiased account of the research performed (Microba Laboratories Accreditation No. 20563).
Figures


Similar articles
-
Clinical Evaluation of Metagenomic Next-Generation Sequencing Method for the Diagnosis of Suspected Ascitic Infection in Patients with Liver Cirrhosis in a Clinical Laboratory.Microbiol Spectr. 2023 Feb 14;11(1):e0294622. doi: 10.1128/spectrum.02946-22. Epub 2023 Jan 10. Microbiol Spectr. 2023. PMID: 36625589 Free PMC article.
-
Augmented pathogen detection in brain abscess using metagenomic next-generation sequencing: a retrospective cohort study.Microbiol Spectr. 2024 Oct 3;12(10):e0032524. doi: 10.1128/spectrum.00325-24. Epub 2024 Sep 12. Microbiol Spectr. 2024. PMID: 39264158 Free PMC article.
-
Metagenomic next-generation sequencing of osteoarticular tissue for the diagnosis of suspected osteoarticular tuberculosis.Microbiol Spectr. 2024 Nov 8;12(12):e0359823. doi: 10.1128/spectrum.03598-23. Online ahead of print. Microbiol Spectr. 2024. PMID: 39513695 Free PMC article.
-
Application of metagenomic next-generation sequencing in the diagnosis of infectious diseases.Front Cell Infect Microbiol. 2024 Nov 15;14:1458316. doi: 10.3389/fcimb.2024.1458316. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39619659 Free PMC article. Review.
-
Clinical Metagenomic Next-Generation Sequencing for Diagnosis of Central Nervous System Infections: Advances and Challenges.Mol Diagn Ther. 2024 Sep;28(5):513-523. doi: 10.1007/s40291-024-00727-9. Epub 2024 Jul 11. Mol Diagn Ther. 2024. PMID: 38992308 Review.
Cited by
-
Advancing metagenomic classification with NABAS+: a novel alignment-based approach.NAR Genom Bioinform. 2025 Jul 4;7(3):lqaf092. doi: 10.1093/nargab/lqaf092. eCollection 2025 Sep. NAR Genom Bioinform. 2025. PMID: 40630933 Free PMC article.
References
-
- Brendish NJ, Beard KR, Malachira AK, Tanner AR, Sanga-Nyirongo L, Gwiggner M, et al. Clinical impact of syndromic molecular point-of-care testing for gastrointestinal pathogens in adults hospitalised with suspected gastroenteritis (GastroPOC): a pragmatic, open-label, randomised controlled trial. Lancet Infect Dis. 2023;23(8):945–55. - PubMed
-
- Ozyurt OK, Saglik I, Ozhak B, Mutlu D, Levent B, Donmez L, et al. Evaluation of Diagnostic Performance of BD Max EBP Assay in Patients with Diarrheal Illness. In Review; 2021. https://www.researchsquare.com/article/rs-258584/v1. Accessed 6 Jan 2022.
LinkOut - more resources
Full Text Sources
Miscellaneous

