Alterations in genes associated with cytosolic RNA sensing in whole blood are associated with coronary microvascular disease in SLE
- PMID: 39828802
- PMCID: PMC11743764
- DOI: 10.1038/s41598-024-82190-4
Alterations in genes associated with cytosolic RNA sensing in whole blood are associated with coronary microvascular disease in SLE
Abstract
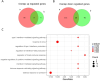
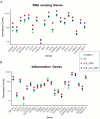
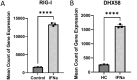
Systemic lupus erythematosus (SLE) patients are 90% women and over three times more likely to die of cardiovascular disease than women in the general population. Chest pain with no obstructive cardiac disease is associated with coronary microvascular disease (CMD), where narrowing of the small blood vessels can lead to ischemia, and frequently reported by SLE patients. Using whole blood RNA samples, we asked whether gene signatures discriminate SLE patients with coronary microvascular dysfunction (CMD) on cardiac MRI (n = 4) from those without (n = 7) and whether any signaling pathway is linked to the underlying pathobiology of SLE CMD. RNA-seq analysis revealed 143 differentially expressed (DE) genes between the SLE and healthy control (HC) groups, with virus defense and interferon (IFN) signaling being the key pathways identified as enriched in SLE as expected. We next conducted a comparative analysis of genes differentially expressed in SLE-CMD and SLE-non-CMD relative to HC samples. Our analysis highlighted differences in IFN signaling, RNA sensing and ADP-ribosylation pathways between SLE-CMD and SLE-non-CMD. This is the first study to investigate possible gene signatures associating with CMD in SLE, and our data strongly suggests that distinct molecular mechanisms underly vascular changes in CMD and non-CMD involvement in SLE.
Keywords: Coronary microvascular disease; Gene expression; Interferon; Systemic lupus erythematosus.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: The experiments were approved by the institutional review board at Cedars-Sinai Medical Center, which oversees the approval of experiments, specifically in the realm of medical ethics. We confirm that all experiments were conducted in accordance with relevant guidelines and regulations, including the Principles of Human Experimentation Ethics and Good Clinical Practice.
Figures


Update of
-
Alterations in genes associated with cytosolic RNA sensing in whole blood are associated with coronary microvascular disease in SLE.Res Sq [Preprint]. 2024 Apr 26:rs.3.rs-4171759. doi: 10.21203/rs.3.rs-4171759/v1. Res Sq. 2024. Update in: Sci Rep. 2025 Jan 19;15(1):2457. doi: 10.1038/s41598-024-82190-4. PMID: 38746373 Free PMC article. Updated. Preprint.
Similar articles
-
Alterations in genes associated with cytosolic RNA sensing in whole blood are associated with coronary microvascular disease in SLE.Res Sq [Preprint]. 2024 Apr 26:rs.3.rs-4171759. doi: 10.21203/rs.3.rs-4171759/v1. Res Sq. 2024. Update in: Sci Rep. 2025 Jan 19;15(1):2457. doi: 10.1038/s41598-024-82190-4. PMID: 38746373 Free PMC article. Updated. Preprint.
-
Coronary Microvascular Dysfunction in Systemic Lupus Erythematosus.J Am Heart Assoc. 2021 Jul 6;10(13):e018555. doi: 10.1161/JAHA.120.018555. Epub 2021 Jun 16. J Am Heart Assoc. 2021. PMID: 34132099 Free PMC article.
-
Disentangling the riddle of systemic lupus erythematosus with antiphospholipid syndrome: blood transcriptome analysis reveals a less-pronounced IFN-signature and distinct molecular profiles in venous versus arterial events.Ann Rheum Dis. 2024 Aug 27;83(9):1132-1143. doi: 10.1136/ard-2024-225664. Ann Rheum Dis. 2024. PMID: 38609158 Free PMC article.
-
Coronary Microvascular Dysfunction in Patients With Systemic Lupus Erythematosus and Chest Pain.Front Cardiovasc Med. 2022 Apr 15;9:867155. doi: 10.3389/fcvm.2022.867155. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35498009 Free PMC article. Review.
-
Coronary Microvascular Dysfunction and Estrogen Receptor Signaling.Trends Endocrinol Metab. 2020 Mar;31(3):228-238. doi: 10.1016/j.tem.2019.11.001. Epub 2019 Nov 29. Trends Endocrinol Metab. 2020. PMID: 31787492 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- U01 HL649241/HL/NHLBI NIH HHS/United States
- N01 HV068161/HV/NHLBI NIH HHS/United States
- 1R03AG032631/AG/NIA NIH HHS/United States
- N01 HV068163/HL/NHLBI NIH HHS/United States
- R03 AG032631/AG/NIA NIH HHS/United States
- N01-HV-68164/HL/NHLBI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- N01-HV-68162/HL/NHLBI NIH HHS/United States
- U01 HL649141/HL/NHLBI NIH HHS/United States
- UL1TR000124/TR/NCATS NIH HHS/United States
- N01-HV-68161/HL/NHLBI NIH HHS/United States
- N01 HV068164/HV/NHLBI NIH HHS/United States
- MO1-RR0042/RR/NCRR NIH HHS/United States
- R01 HL153500/HL/NHLBI NIH HHS/United States
- M01 RR000425/RR/NCRR NIH HHS/United States
- M01 RR000042/RR/NCRR NIH HHS/United States
- N01 HV068162/HL/NHLBI NIH HHS/United States
- U0164829/HL/NHLBI NIH HHS/United States
- R01 AI164504/AI/NIAID NIH HHS/United States
- R01 HL146158/HL/NHLBI NIH HHS/United States
- N01-HV-68163/HL/NHLBI NIH HHS/United States
- 1R01AI164504/National Institute of Allergy and Infectious Diseases
- W81XWH-18-1-0709/Congressionally Directed Medical Research Programs
- R01 HL090957/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

