Evidence for peroxisomal redundancy among the glucose-6-phosphate dehydrogenase isoforms of Arabidopsis thaliana
- PMID: 39829315
- PMCID: PMC12125578
- DOI: 10.1093/pcp/pcaf012
Evidence for peroxisomal redundancy among the glucose-6-phosphate dehydrogenase isoforms of Arabidopsis thaliana
Abstract
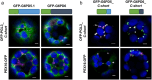
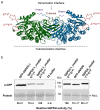
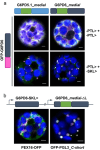
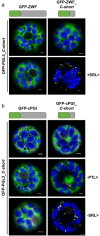
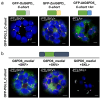
The oxidative pentose phosphate pathway (OPPP) plays an important role in the generation of reducing power in all eukaryotes. In plant cells, the OPPP operates in several cellular compartments, but as full cycle only in the plastid stroma where it is essential. As suggested by our recent results, OPPP reactions are also mandatory inside peroxisomes, at least during fertilization. For the first enzyme of the OPPP, glucose-6-phosphate dehydrogenase (G6PD), we previously showed that one Arabidopsis isoform (G6PD1) can be directed to peroxisomes under specific circumstances. Since g6pd1 knock-out plants are viable, we aimed at elucidating potential redundancy regarding peroxisomal targeting among the other G6PD isoforms. Localization studies of so far cytosolic annotated G6PD5 and G6PD6 (both ending -PTL>) using different reporter fusions of full-length versus the last 50 amino acids revealed that GFP-C-short versions are efficiently imported into peroxisomes. Modification of the final tripeptide to a canonical peroxisomal targeting signal type 1 (PTS1) also resulted in peroxisomal localization of the full-length versions and revealed that G6PD5/6 import may occur as homo- or heterodimer. Interestingly, the new noncanonical PTS1 motif is highly conserved among the cytosolic G6PD isoforms of the Angiosperms, whereas members of the Poaceae (rice and maize) possess two variants, one ending with an additional amino acid (-PTLA>) and the other one extended by a stronger PTS1 motif. From both evolutionary and physiological perspectives, we postulate that G6PD import as homo- and heterodimer restricted the acquisition of more efficient peroxisomal targeting motifs to leave some G6PDH activity in the cytosol.
Keywords: G6PD; OPPP; dimeric protein import; dual targeting; peroxisomes; targeting motifs.
© The Author(s) 2025. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists.
Conflict of interest statement
None declared.
Figures



Similar articles
-
Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol.Plant J. 2011 Jun;66(5):745-58. doi: 10.1111/j.1365-313X.2011.04535.x. Epub 2011 Mar 21. Plant J. 2011. PMID: 21309870
-
Alternative splicing of Arabidopsis G6PD5 recruits NADPH-producing OPPP reactions to the endoplasmic reticulum.Front Plant Sci. 2022 Sep 2;13:909624. doi: 10.3389/fpls.2022.909624. eCollection 2022. Front Plant Sci. 2022. PMID: 36119606 Free PMC article.
-
Evidence for dual targeting control of Arabidopsis 6-phosphogluconate dehydrogenase isoforms by N-terminal phosphorylation.J Exp Bot. 2024 May 20;75(10):2848-2866. doi: 10.1093/jxb/erae077. J Exp Bot. 2024. PMID: 38412416 Free PMC article.
-
Prediction of Peroxisomal Matrix Proteins in Plants.Subcell Biochem. 2018;89:125-138. doi: 10.1007/978-981-13-2233-4_5. Subcell Biochem. 2018. PMID: 30378021 Review.
-
Peroxisome protein import: some answers, more questions.Curr Opin Plant Biol. 2005 Dec;8(6):640-7. doi: 10.1016/j.pbi.2005.09.009. Epub 2005 Sep 22. Curr Opin Plant Biol. 2005. PMID: 16182600 Review.
References
-
- Apanasets O., Grou C.P., Van Veldhoven P.P., Brees C., Wang B., Nordgren M., et al. (2014) PEX5, the shuttling import receptor for peroxisomal matrix proteins, is a redox-sensitive protein: PTS1 protein import and oxidative stress. Traffic 15: 94–103. - PubMed
-
- Buchanan B.B. (1991) Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Arch. Biochem. Biophys. 288: 1–9. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

