EAVLD 2024 - 7th Congress of the European Association of Veterinary Laboratory Diagnosticians
- PMID: 39829721
- PMCID: PMC11740014
- DOI: 10.4081/ijfs.2024.13488
EAVLD 2024 - 7th Congress of the European Association of Veterinary Laboratory Diagnosticians
Abstract
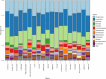
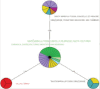
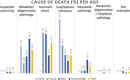
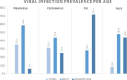
This abstract book contains the abstracts presented at the 7th Congress of the European Association of Veterinary Laboratory Diagnosticians.
Copyright © 2024, the Author(s).
Figures































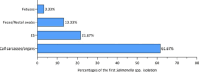




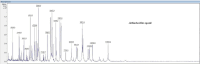
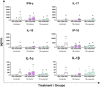
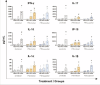






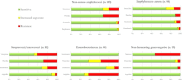

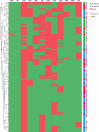


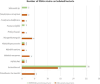
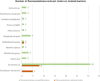







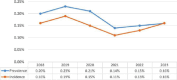
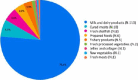

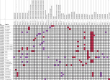






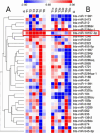
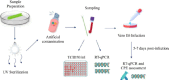
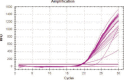



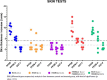

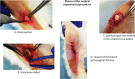

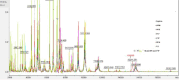
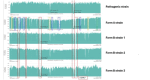


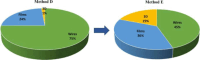



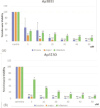
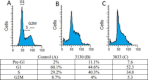
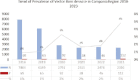
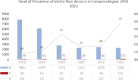


Similar articles
-
Abstracts of the European Society of Veterinary Clinical Pathology (ESVCP) 10th Annual Congress in conjunction with the International Society for Animal Clinical Pathology (ISACP) 13th Biennial Congress, the Association for Acute Phase Proteins (APP) 7th Congress, and the Association for European Comparative Clinical Pathology (AECCP) 8th Congress. Barcelona, Spain. September 30–October 3, 2008.Vet Clin Pathol. 2008 Dec;37 Suppl 1:19-40. doi: 10.1111/j.1939-165X.2008.00097.x. Vet Clin Pathol. 2008. PMID: 19068069 No abstract available.
-
The Scientific Value of Abstracts on Prostate Cancer Presented at the European Association of Urology Congresses.Front Surg. 2021 Jun 15;8:683359. doi: 10.3389/fsurg.2021.683359. eCollection 2021. Front Surg. 2021. PMID: 34212000 Free PMC article.
-
What is the fate of scientific abstracts? The publication rates of abstracts presented at the 7th National Congress of Gynecology and Obstetrics.Turk J Obstet Gynecol. 2015 Mar;12(1):25-29. doi: 10.4274/tjod.77785. Epub 2015 Mar 15. Turk J Obstet Gynecol. 2015. PMID: 28913036 Free PMC article.
-
Publication rate of abstracts presented at the Congress of the European Federation of National Associations of Orthopaedics and Traumatology (EFORT).Orthop Traumatol Surg Res. 2019 Oct 3:1453-1457. doi: 10.1016/j.otsr.2019.07.014. Online ahead of print. Orthop Traumatol Surg Res. 2019. PMID: 31588034
-
Highlights of the 30th Annual Congress of the EANM, Vienna 2017: "Yes we can - make nuclear medicine great again".Eur J Nucl Med Mol Imaging. 2018 Sep;45(10):1781-1794. doi: 10.1007/s00259-018-4029-9. Epub 2018 May 3. Eur J Nucl Med Mol Imaging. 2018. PMID: 29725717 Free PMC article. Review.
References
-
- Simner PJ, Rauch CA, Martin IW, Sullivan KV, Rhoads D, Rolf R, She R, Souers RJ, Wojewoda C, Humphries RM. 2022. Raising the bar: improving antimicrobial resistance detection by clinical laboratories by ensuring use of current breakpoints. Open Forum Infect Dis 9. 10.1093/ofid/ofac007 - DOI - PMC - PubMed
-
- Rossen JWA, Friedrich AW, Moran-Gilad J, ESCMID Study Group for Genomic and Molecular Diagnostics (ESGMD) . 2018. Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin Microbiol Infect 24:355–360. Crossref. PubMed. ISI. - PubMed
-
- Timofte D., Broens E. M., Guardabassi L., Pomba C., Allerton F., Ikonomopoulos J., . . . European Network for Optimisation of Veterinary Antimicrobial Treatment (ENOVAT); ESCMID Study Group for Veterinary Microbiology (ESGVM); European College of Veterinary Microbiology (ECVM) and European Association of Veterinary Laboratory Diagnosticians (EAVLD) (2021). Driving laboratory standardisation of bacterial culture and antimicrobial susceptibility testing in veterinary clinical microbiology in Europe and beyond. Journal of Clinical Microbiology. 59, doi:10.1128/jcm.02572-20 - PMC - PubMed
-
- Koritnik Tom, Cvetkovikj Iskra, Zendri Flavia, Blum Shlomo Eduardo, Chaintoutis Serafeim Christos, Kopp Peter A., Hare Cassia, Štritof Zrinka, Kittl Sonja, Gonçalves José, Zdovc Irena, Paulshus Erik, Laconi Andrea, Singleton David, Allerton Fergus, Broens Els M., Damborg Peter, Timofte Dorina. Towards harmonised laboratory methodologies in veterinary clinical bacteriologyoutcomes of a European survey. Frontiers in Microbiology, under review (Manuscript ID: 1443755) - PMC - PubMed
-
- Marques C, Gama LT, Belas A, Bergström K, Beurlet S, Briend-Marchal A, Broens EM, Costa M, Criel D, Damborg P, van Dijk MAM, van Dongen AM, Dorsch R, Espada CM, Gerber B, Kritsepi-Konstantinou M, Loncaric I, Mion D, Misic D, Movilla R, Overesch G, Perreten V, Roura X, Steenbergen J, Timofte D, Wolf G, Zanoni RG, Schmitt S, Guardabassi L, Pomba C. 2016. European multicenter study on antimicrobial resistance in bacteria isolated from companion animal urinary tract infections. BMC Vet Res 12:213. doi: 10.1186/s12917-016-0840- - PMC - PubMed
LinkOut - more resources
Full Text Sources

