This is a preprint.
Single-cell profiling of trabecular meshwork identifies mitochondrial dysfunction in a glaucoma model that is protected by vitamin B3 treatment
- PMID: 39829808
- PMCID: PMC11741249
- DOI: 10.1101/2024.11.01.621152
Single-cell profiling of trabecular meshwork identifies mitochondrial dysfunction in a glaucoma model that is protected by vitamin B3 treatment
Abstract
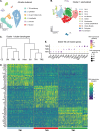
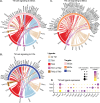
Since the trabecular meshwork (TM) is central to intraocular pressure (IOP) regulation and glaucoma, a deeper understanding of its genomic landscape is needed. We present a multimodal, single-cell resolution analysis of mouse limbal cells (includes TM). In total, we sequenced 9,394 wild-type TM cell transcriptomes. We discovered three TM cell subtypes with characteristic signature genes validated by immunofluorescence on tissue sections and whole-mounts. The subtypes are robust, being detected in datasets for two diverse mouse strains and in independent data from two institutions. Results show compartmentalized enrichment of critical pathways in specific TM cell subtypes. Distinctive signatures include increased expression of genes responsible for 1) extracellular matrix structure and metabolism (TM1 subtype), 2) secreted ligand signaling to support Schlemm's canal cells (TM2), and 3) contractile and mitochondrial/metabolic activity (TM3). ATAC-sequencing data identified active transcription factors in TM cells, including LMX1B. Mutations in LMX1B cause high IOP and glaucoma. LMX1B is emerging as a key transcription factor for normal mitochondrial function and its expression is much higher in TM3 cells than other limbal cells. To understand the role of LMX1B in TM function and glaucoma, we single-cell sequenced limbal cells from Lmx1b V265D/+ mutant mice (2,491 TM cells). In V265D/+ mice, TM3 cells were uniquely affected by pronounced mitochondrial pathway changes. Mitochondria in TM cells of V265D/+ mice are swollen with a reduced cristae area, further supporting a role for mitochondrial dysfunction in the initiation of IOP elevation in these mice. Importantly, treatment with vitamin B3 (nicotinamide), to enhance mitochondrial function and metabolic resilience, significantly protected Lmx1b mutant mice from IOP elevation.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
