This is a preprint.
Fast and flexible minimizer digestion with digest
- PMID: 39829865
- PMCID: PMC11741343
- DOI: 10.1101/2025.01.02.631161
Fast and flexible minimizer digestion with digest
Update in
-
Fast and flexible minimizer digestion with digest.Bioinformatics. 2025 Jul 1;41(7):btaf368. doi: 10.1093/bioinformatics/btaf368. Bioinformatics. 2025. PMID: 40581603 Free PMC article.
Abstract
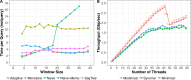
Minimizer digestion is an increasingly common component of bioinformatics tools, including tools for De Bruijn-Graph assembly and sequence classification. We describe a new open source tool and library to facilitate efficient digestion of genomic sequences. It can produce digests based on the related ideas of minimizers, modimizers or syncmers. Digest uses efficient data structures, scales well to many threads, and produces digests with expected spacings between digested elements. Digest is implemented in C++17 with a Python API, and is available open-source at https://github.com/VeryAmazed/digest.
Keywords: digestion; minimizers; sequence analysis; syncmers.
Conflict of interest statement
Competing interests All authors contributed to and reviewed the manuscript. No competing interest is declared.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
