This is a preprint.
Multi-omics and biochemical reconstitution reveal CDK7-dependent mechanisms controlling RNA polymerase II function at gene 5'- and 3'-ends
- PMID: 39829884
- PMCID: PMC11741307
- DOI: 10.1101/2025.01.08.632016
Multi-omics and biochemical reconstitution reveal CDK7-dependent mechanisms controlling RNA polymerase II function at gene 5'- and 3'-ends
Update in
-
Multi-omics and biochemical reconstitution reveal CDK7-dependent mechanisms controlling RNA polymerase II function at gene 5'- and 3' ends.Cell Rep. 2025 Jul 22;44(7):115904. doi: 10.1016/j.celrep.2025.115904. Epub 2025 Jun 25. Cell Rep. 2025. PMID: 40570372 Free PMC article.
Abstract
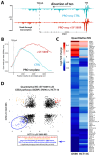
CDK7 regulates RNA polymerase II (RNAPII) initiation, elongation, and termination through incompletely understood mechanisms. Because contaminating kinases precluded CDK7 analysis with nuclear extracts, we completed biochemical assays with purified factors. Reconstitution of RNAPII transcription initiation showed CDK7 inhibition slowed and/or paused RNAPII promoter-proximal transcription, which reduced re-initiation. These CDK7-regulatory functions were Mediator- and TFIID-dependent. Similarly in human cells, CDK7 inhibition reduced transcription by suppressing RNAPII activity at promoters, consistent with reduced initiation and/or re-initiation. Moreover, widespread 3'-end readthrough transcription was observed in CDK7-inhibited cells; mechanistically, this occurred through rapid nuclear depletion of RNAPII elongation and termination factors, including high-confidence CDK7 targets. Collectively, these results define how CDK7 governs RNAPII function at gene 5'-ends and 3'-ends, and reveal that nuclear abundance of elongation and termination factors is kinase-dependent. Because 3'-readthrough transcription is commonly induced during stress, our results further suggest regulated suppression of CDK7 activity may enable this RNAPII transcriptional response.
Conflict of interest statement
DJT received some funding support for this project from Syros Pharmaceuticals, Inc. R.D.D. is a founder of Arpeggio Biosciences. The remaining authors declare no competing interests.
Figures






References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
