This is a preprint.
Neural plate pre-patterning enables specification of intermediate neural progenitors in the spinal cord
- PMID: 39829904
- PMCID: PMC11741283
- DOI: 10.1101/2025.01.09.632276
Neural plate pre-patterning enables specification of intermediate neural progenitors in the spinal cord
Abstract
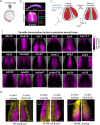
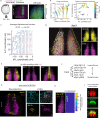
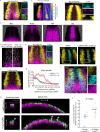
Dorsal-ventral patterning of neural progenitors in the posterior neural tube, which gives rise to the spinal cord, has served as a model system to understand how extracellular signals organize developing tissues. While previous work has shown that signaling gradients diversify progenitor fates at the dorsal and ventral ends of the tissue, the basis of fate specification in intermediate regions has remained unclear. Here we use zebrafish to investigate the neural plate, which precedes neural tube formation, and show that its pre-patterning by a distinct signaling environment enables intermediate fate specification. Systematic spatial analysis of transcription factor (TF) expression and signaling activity using a reference-based mapping approach shows that the neural plate is partitioned into a striking complexity of TF co-expression states that, in part, correspond to the activity of gastrulation signals such as FGF and Wnt that persist through axis extension. Using in toto analysis of cellular movement combined with fate mapping, we find that dbx1b-expressing intermediate progenitors (p0) originate from a neural-plate specific state characterized by transient co-expression of the TFs pax3a, olig4 and her3. Finally, we show that this state is defined by Wnt signaling in the posterior neural plate and that ectopic Wnt activation within pax3a/olig4+ cells is sufficient to promote dbx1b expression. Our data broadly support a model in which neural progenitor specification occurs through the sequential use of multiple signals to progressively diversify the neural tissue as it develops. This has implications for in vitro differentiation of spinal cord cell types and for understanding signal-based patterning in other developmental contexts.
Figures

Similar articles
-
Wnt signaling regulates neural plate patterning in distinct temporal phases with dynamic transcriptional outputs.Dev Biol. 2020 Jun 15;462(2):152-164. doi: 10.1016/j.ydbio.2020.03.016. Epub 2020 Mar 31. Dev Biol. 2020. PMID: 32243887 Free PMC article.
-
Fate Specification of Neural Plate Border by Canonical Wnt Signaling and Grhl3 is Crucial for Neural Tube Closure.EBioMedicine. 2015 Apr 18;2(6):513-27. doi: 10.1016/j.ebiom.2015.04.012. eCollection 2015 Jun. EBioMedicine. 2015. PMID: 26288816 Free PMC article.
-
BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border.Development. 2012 Nov;139(22):4220-31. doi: 10.1242/dev.081497. Epub 2012 Oct 3. Development. 2012. PMID: 23034628 Free PMC article.
-
Morphogens and the control of cell proliferation and patterning in the spinal cord.Cell Cycle. 2007 Nov 1;6(21):2640-9. doi: 10.4161/cc.6.21.4822. Epub 2007 Aug 1. Cell Cycle. 2007. PMID: 17912034 Review.
-
Neural patterning in the vertebrate embryo.Int Rev Cytol. 2001;203:447-82. doi: 10.1016/s0074-7696(01)03013-3. Int Rev Cytol. 2001. PMID: 11131523 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
