Targeting intracellular cancer proteins with tumor-microenvironment-responsive bispecific nanobody-PROTACs for enhanced therapeutic efficacy
- PMID: 39830023
- PMCID: PMC11742431
- DOI: 10.1002/mco2.70068
Targeting intracellular cancer proteins with tumor-microenvironment-responsive bispecific nanobody-PROTACs for enhanced therapeutic efficacy
Abstract
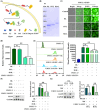
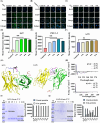
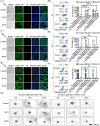
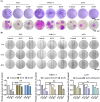
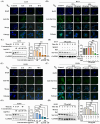
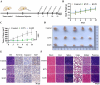
Proteolysis targeting chimeras (PROTACs) are pivotal in cancer therapy for their ability to degrade specific proteins. However, their non-specificity can lead to systemic toxicity due to protein degradation in normal cells. To address this, we have integrated a nanobody into the PROTACs framework and leveraged the tumor microenvironment to enhance drug specificity. In this study, we engineered BumPeD, a novel bispecific nanobody-targeted PROTACs-like platform, by fusing two nanobodies with a Furin protease cleavage site (RVRR) and a degron sequence (ALAPYIP or KIGLGRQKPPKATK), enabling the tumor microenvironment to direct the degradation of intracellular proteins. We utilized KN035 and Nb4A to target PD-L1 (programmed death ligand 1) on the cell surface and intracellular Survivin, respectively. In vitro experiments showed that BumPeD triggers Survivin degradation via the ubiquitin-proteasome pathway, inducing tumor apoptosis and suppressing bladder tumor cell proliferation and migration. In vivo experiments further confirmed BumPeD's robust anti-tumor efficacy, underscoring its potential as a precise protein degradation strategy for cancer therapy. Our platform provides a systematic approach to developing effective and practical protein degraders, offering a targeted theoretical basis and experimental support for the development of novel degradative drugs, as well as new directions for cancer therapy.
Keywords: PD‐L1 and Survivin; nanobody; proteolysis targeting chimeras (PROTACs); targeted degradation; tumor microenvironment.
© 2025 The Author(s). MedComm published by Sichuan International Medical Exchange & Promotion Association (SCIMEA) and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures

Similar articles
-
Self-Assembled Peptide-Derived Proteolysis-Targeting Chimera (PROTAC) Nanoparticles for Tumor-Targeted and Durable PD-L1 Degradation in Cancer Immunotherapy.Angew Chem Int Ed Engl. 2025 Jan 27;64(5):e202414146. doi: 10.1002/anie.202414146. Epub 2024 Dec 17. Angew Chem Int Ed Engl. 2025. PMID: 39572518
-
Enzyme-Activated Orthogonal Proteolysis Chimeras for Tumor Microenvironment-Responsive Immunomodulation.Angew Chem Int Ed Engl. 2025 May 26;64(22):e202423057. doi: 10.1002/anie.202423057. Epub 2025 Feb 19. Angew Chem Int Ed Engl. 2025. PMID: 39932237
-
Powering up targeted protein degradation through active and passive tumour-targeting strategies: Current and future scopes.Pharmacol Ther. 2024 Nov;263:108725. doi: 10.1016/j.pharmthera.2024.108725. Epub 2024 Sep 24. Pharmacol Ther. 2024. PMID: 39322067 Review.
-
Split-Cas9-based targeted gene editing and nanobody-mediated proteolysis-targeting chimeras optogenetically coordinated regulation of Survivin to control the fate of cancer cells.Clin Transl Med. 2023 Aug;13(8):e1382. doi: 10.1002/ctm2.1382. Clin Transl Med. 2023. PMID: 37620295 Free PMC article.
-
Precise Modulation of Protein Degradation by Smart PROTACs.Chembiochem. 2025 Jan 2;26(1):e202400682. doi: 10.1002/cbic.202400682. Epub 2024 Nov 11. Chembiochem. 2025. PMID: 39367518 Review.
Cited by
-
Nanobodies and their derivatives: pioneering the future of cancer immunotherapy.Cell Commun Signal. 2025 Jun 5;23(1):271. doi: 10.1186/s12964-025-02270-4. Cell Commun Signal. 2025. PMID: 40474230 Free PMC article. Review.
-
Immune Memory: A New Frontier in Treating Recurrent Inflammatory Skin Diseases.Clin Rev Allergy Immunol. 2025 Mar 18;68(1):31. doi: 10.1007/s12016-025-09039-0. Clin Rev Allergy Immunol. 2025. PMID: 40100550 Review.
-
The unexplored mechanism of antitumoral effect of pirfenidone in melanoma cells.Sci Rep. 2025 Aug 1;15(1):28071. doi: 10.1038/s41598-025-13584-1. Sci Rep. 2025. PMID: 40751072 Free PMC article.
References
-
- He S, Dong G, Cheng J, Wu Y, Sheng C. Strategies for designing proteolysis targeting chimaeras (PROTACs). Med Res Rev. 2022;42(3):1280‐1342. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
