This is a preprint.
Deleterious coding variation associated with autism is consistent across populations, as exemplified by admixed Latin American populations
- PMID: 39830258
- PMCID: PMC11741445
- DOI: 10.1101/2024.12.27.24319460
Deleterious coding variation associated with autism is consistent across populations, as exemplified by admixed Latin American populations
Abstract
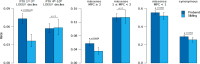
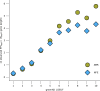
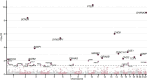
The past decade has seen remarkable progress in identifying genes that, when impacted by deleterious coding variation, confer high risk for autism spectrum disorder (ASD), intellectual disability, and other developmental disorders. However, most underlying gene discovery efforts have focused on individuals of European ancestry, limiting insights into genetic risks across diverse populations. To help address this, the Genomics of Autism in Latin American Ancestries Consortium (GALA) was formed, presenting here the largest sequencing study of ASD in Latin American individuals (n>15,000). We identified 35 genome-wide significant (FDR < 0.05) ASD risk genes, with substantial overlap with findings from European cohorts, and highly constrained genes showing consistent signal across populations. The results provide support for emerging (e.g., MARK2, YWHAG, PACS1, RERE, SPEN, GSE1, GLS, TNPO3, ANKRD17) and established ASD genes, and for the utility of genetic testing approaches for deleterious variants in diverse populations, while also demonstrating the ongoing need for more inclusive genetic research and testing. We conclude that the biology of ASD is universal and not impacted to any detectable degree by ancestry.
Conflict of interest statement
Competing interests Lilia Albores declares to be the main author of the CRIDI-ASD interview, she is a professor of the training course for the mentioned instrument, and receives payment for the training.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
