An integrated approach for the accurate detection of HERV-K HML-2 transcription and protein synthesis
- PMID: 39831303
- PMCID: PMC11744191
- DOI: 10.1093/nar/gkaf011
An integrated approach for the accurate detection of HERV-K HML-2 transcription and protein synthesis
Abstract
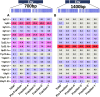
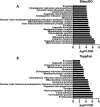
Human endogenous retroviruses (HERVs) occupy a large portion of the human genome. Most HERVs are transcriptionally silent, but they can be reactivated during pathological states such as viral infection and certain cancers. The HERV-K HML-2 clade includes elements that recently integrated have in the human germ line and often contain intact open reading frames that possibly support peptide and protein expression. Understanding HERV-K-host interactions and their potential as biomarkers is problematic due to the high similarity among different elements. Previously, we described a long-read single molecule real-time sequencing (PacBio) strategy to analyze HERV-K RNA expression profiles in different cell types. However, identifying HERV-K HML-2 proteins accurately is difficult without robust and reliable methods and reagents. Here we present a new approach to characterize the HML-2 elements that (a) are being translated and (b) produce enough protein to be detected and identified by mass spectrometry. Our data reveal that RNA expression profiling alone cannot accurately predict which HML-2 elements are responsible for protein production, as we observe several differences between the highest expressed RNAs and the elements that are the predominant source of HERV-K HML-2 protein synthesis. These studies represent an important advance toward untangling the complexity of HERV-K-host interactions.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
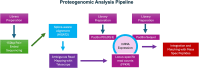
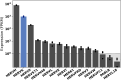
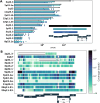
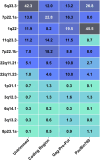

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

