Two complementing in vivo selection systems based on CCA-trimming exonucleases as a tool to monitor, select and evaluate enzymatic features of tRNA nucleotidyltransferases
- PMID: 39831457
- PMCID: PMC11784652
- DOI: 10.1080/15476286.2025.2453963
Two complementing in vivo selection systems based on CCA-trimming exonucleases as a tool to monitor, select and evaluate enzymatic features of tRNA nucleotidyltransferases
Abstract
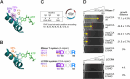
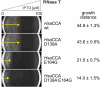
tRNA nucleotidyltransferase represents a ubiquitous and essential activity that adds the indispensable CCA triplet to the 3'-end of tRNAs. To fulfill this function, the enzyme contains a set of highly conserved motifs whose coordinated interplay is crucial for the sequence-specific CCA polymerization. In the human enzyme, alterations within these regions have been shown to lead to the manifestation of disease. Recently, we developed an in vivo screening system that allows for the selection and analysis of tRNA nucleotidyltransferase variants by challenging terminal AMP incorporation into tRNA during induced RNase T-catalyzed CCA-decay. Here, we extend this method for screening of full CCA-end repair by utilizing the CCA-trimming activity of exonuclease LCCR4. To demonstrate the combined potential of these two in vivo selection systems, we applied a semi-rational library design to investigate the mode of operation of catalytically important motifs in the human CCA-adding enzyme. This approach revealed unexpected requirements for amino acid composition in two motifs and gives new insights into the mechanism of CCA addition. The data show the potential of these RNase-based screening systems, as they allow the detection of enzyme variations that would not have been identified by a conventional rational approach. Furthermore, the combination of both RNase T and LCCR4 systems can be used to investigate and dissect the effects of pathogenic mutations on C- and A-addition.
Keywords: CCA-adding enzyme; CCA-trimming exonuclease; RNase LCCR4; RNase T; in vivo screening of CCA-adding activities; tRNA nucleotidyltransferase.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures







Similar articles
-
Examining tRNA 3'-ends in Escherichia coli: teamwork between CCA-adding enzyme, RNase T, and RNase R.RNA. 2018 Mar;24(3):361-370. doi: 10.1261/rna.064436.117. Epub 2017 Nov 27. RNA. 2018. PMID: 29180590 Free PMC article.
-
Domain movements during CCA-addition: a new function for motif C in the catalytic core of the human tRNA nucleotidyltransferases.RNA Biol. 2015;12(4):435-46. doi: 10.1080/15476286.2015.1018502. RNA Biol. 2015. PMID: 25849199 Free PMC article.
-
A single catalytically active subunit in the multimeric Sulfolobus shibatae CCA-adding enzyme can carry out all three steps of CCA addition.J Biol Chem. 2004 Sep 17;279(38):40130-6. doi: 10.1074/jbc.M405518200. Epub 2004 Jul 19. J Biol Chem. 2004. PMID: 15265870
-
A tRNA's fate is decided at its 3' end: Collaborative actions of CCA-adding enzyme and RNases involved in tRNA processing and degradation.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):433-441. doi: 10.1016/j.bbagrm.2018.01.012. Epub 2018 Jan 31. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29374586 Review.
-
The CCA-adding enzyme: A central scrutinizer in tRNA quality control.Bioessays. 2015 Sep;37(9):975-82. doi: 10.1002/bies.201500043. Epub 2015 Jul 14. Bioessays. 2015. PMID: 26172425 Review.
Cited by
-
Armless hairpin-like tRNAs in Romanomermis culicivorax: Evolutionary adaptation of a mitochondrial elongation factor EF-Tu.J Biol Chem. 2025 Jul;301(7):110294. doi: 10.1016/j.jbc.2025.110294. Epub 2025 May 24. J Biol Chem. 2025. PMID: 40419126 Free PMC article.
References
-
- Sprinzl M, Cramer F.. The -C-C-A end of tRNA and its role in protein biosynthesis. Prog Nucleic Acid Res Mol Biol. 1979;22:1–69. - PubMed
-
- Aebi M, Kirchner G, Chen JY, et al. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990;265(27):16216–16220. doi: 10.1016/S0021-9258(17)46210-7 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
