Sir2 and Fun30 regulate ribosomal DNA replication timing via MCM helicase positioning and nucleosome occupancy
- PMID: 39831552
- PMCID: PMC11745493
- DOI: 10.7554/eLife.97438
Sir2 and Fun30 regulate ribosomal DNA replication timing via MCM helicase positioning and nucleosome occupancy
Abstract
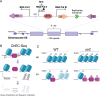
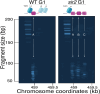
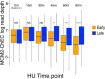
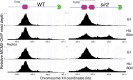
The association between late replication timing and low transcription rates in eukaryotic heterochromatin is well known, yet the specific mechanisms underlying this link remain uncertain. In Saccharomyces cerevisiae, the histone deacetylase Sir2 is required for both transcriptional silencing and late replication at the repetitive ribosomal DNA (rDNA) arrays. We have previously reported that in the absence of SIR2, a de-repressed RNA PolII repositions MCM replicative helicases from their loading site at the ribosomal origin, where they abut well-positioned, high-occupancy nucleosomes, to an adjacent region with lower nucleosome occupancy. By developing a method that can distinguish activation of closely spaced MCM complexes, here we show that the displaced MCMs at rDNA origins have increased firing propensity compared to the nondisplaced MCMs. Furthermore, we found that both activation of the repositioned MCMs and low occupancy of the adjacent nucleosomes critically depend on the chromatin remodeling activity of FUN30. Our study elucidates the mechanism by which Sir2 delays replication timing, and it demonstrates, for the first time, that activation of a specific replication origin in vivo relies on the nucleosome context shaped by a single chromatin remodeler.
Keywords: DNA replication; Fun30; S. cerevisiae; Sir2; chromatin; chromatin remodeling enzyme; chromosomes; gene expression; silencing.
© 2024, Lichauco, Foss et al.
Conflict of interest statement
CL, EF, TG, NA, BL, RA, ET, JM, UL, SM, AB No competing interests declared
Figures













Update of
-
Sir2 and Fun30 regulate ribosomal DNA replication timing via MCM helicase positioning and nucleosome occupancy.bioRxiv [Preprint]. 2024 Oct 28:2024.03.21.586113. doi: 10.1101/2024.03.21.586113. bioRxiv. 2024. Update in: Elife. 2025 Jan 20;13:RP97438. doi: 10.7554/eLife.97438. PMID: 38585982 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

