MYC and HSF1 Cooperate to Drive Sensitivity to Polo-like Kinase 1 Inhibitor Volasertib in High-grade Serous Ovarian Cancer
- PMID: 39831777
- PMCID: PMC11799878
- DOI: 10.1158/2767-9764.CRC-24-0400
MYC and HSF1 Cooperate to Drive Sensitivity to Polo-like Kinase 1 Inhibitor Volasertib in High-grade Serous Ovarian Cancer
Abstract
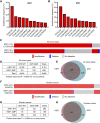
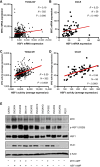
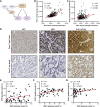
Abstract: Ovarian cancer is a deadly gynecologic disease with frequent recurrence. Current treatments for patients include platinum-based therapy regimens with PARP inhibitors specific for homologous recombination–deficient high-grade serous ovarian cancers (HGSOC). Despite initial effectiveness, patients inevitably develop disease progression as tumor cells acquire resistance. Toward the development of new therapeutic avenues, we describe a gene amplification involving both heat shock factor 1 (HSF1) and MYC, wherein these two genes are co-amplified in more than 30% of patients with HGSOC. We further found that HSF1 and MYC transcriptional activities were highly correlated with human HGSOC tumors and cell lines, suggesting that they may cooperate in the disease. CUT&RUN sequencing for HSF1 and MYC revealed overlapping HSF1 and MYC binding throughout the genome. Moreover, the binding peaks of both transcription factors in HGSOC cells were nearly identical, and a protein–protein interaction between HSF1 and MYC was detected, supporting molecular cooperation. Supporting a functional cooperation of these two transcription factors, the growth of HGSOC cells with the co-amplification was dependent on both HSF1 and MYC. To identify a therapeutic target that could take advantage of this unique HSF1 and MYC dependency, polo-like kinase 1 (PLK1) was correlated with HSF1 and MYC in HGSOC specimens. Targeting PLK1 with volasertib revealed a greater than 200-fold increased potency in HSF1–MYC co-amplified HGSOC cells compared with those with wild-type HSF1 and MYC copy numbers. Although the success of volasertib and other PLK1 inhibitors in clinical trials has been modest, the current study suggests that targeting PLK1 using a precision medicine approach based on HSF1–MYC co-amplification as a biomarker in HGSOC would improve therapy response and patient outcomes.
Significance: We show that HSF1 and MYC genes are co-amplified in more than 30% of HGSOC and demonstrate that HSF1 and MYC functionally cooperate to drive the growth of HGSOC cells. This work provides the foundation for HSF1 and MYC co-amplification as a biomarker for treatment efficacy of the polo-like kinase 1 inhibitor volasertib in HGSOC.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
I. Williams reports grants from Ovarian Cancer Research Alliance during the conduct of the study. R.L. Carpenter reports a patent for 18/418,110 pending. No disclosures were reported by the other authors.
Figures




Update of
-
MYC and HSF1 Cooperate to Drive PLK1 inhibitor Sensitivity in High Grade Serous Ovarian Cancer.bioRxiv [Preprint]. 2024 Jun 13:2024.06.11.598486. doi: 10.1101/2024.06.11.598486. bioRxiv. 2024. Update in: Cancer Res Commun. 2025 Feb 01;5(2):253-266. doi: 10.1158/2767-9764.CRC-24-0400. PMID: 38915574 Free PMC article. Updated. Preprint.
References
-
- ACS . Cancer facts & figures 2024. Atlanta (GA): American Cancer Society; 2024.
-
- Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet 2019;393:1240–53. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

