Twenty years of advances in prediction of nucleic acid-binding residues in protein sequences
- PMID: 39833102
- PMCID: PMC11745544
- DOI: 10.1093/bib/bbaf016
Twenty years of advances in prediction of nucleic acid-binding residues in protein sequences
Abstract
Computational prediction of nucleic acid-binding residues in protein sequences is an active field of research, with over 80 methods that were released in the past 2 decades. We identify and discuss 87 sequence-based predictors that include dozens of recently published methods that are surveyed for the first time. We overview historical progress and examine multiple practical issues that include availability and impact of predictors, key features of their predictive models, and important aspects related to their training and assessment. We observe that the past decade has brought increased use of deep neural networks and protein language models, which contributed to substantial gains in the predictive performance. We also highlight advancements in vital and challenging issues that include cross-predictions between deoxyribonucleic acid (DNA)-binding and ribonucleic acid (RNA)-binding residues and targeting the two distinct sources of binding annotations, structure-based versus intrinsic disorder-based. The methods trained on the structure-annotated interactions tend to perform poorly on the disorder-annotated binding and vice versa, with only a few methods that target and perform well across both annotation types. The cross-predictions are a significant problem, with some predictors of DNA-binding or RNA-binding residues indiscriminately predicting interactions with both nucleic acid types. Moreover, we show that methods with web servers are cited substantially more than tools without implementation or with no longer working implementations, motivating the development and long-term maintenance of the web servers. We close by discussing future research directions that aim to drive further progress in this area.
Keywords: DNA-binding residue; RNA-binding residue; deep learning; intrinsic disorder; machine learning; nucleic acid-binding; protein–DNA interaction; protein–RNA interaction; sequence-based prediction.
© The Author(s) 2025. Published by Oxford University Press.
Figures
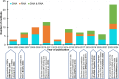
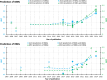
Similar articles
-
Advances in Language-Model-Informed Protein-Nucleic Acid Binding Site Prediction.Methods Mol Biol. 2025;2941:139-151. doi: 10.1007/978-1-0716-4623-6_9. Methods Mol Biol. 2025. PMID: 40601256
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

