Cryo-electron tomography pipeline for plasma membranes
- PMID: 39833141
- PMCID: PMC11747107
- DOI: 10.1038/s41467-025-56045-z
Cryo-electron tomography pipeline for plasma membranes
Abstract
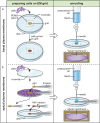
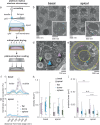
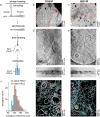
Cryo-electron tomography (cryoET) provides sub-nanometer protein structure within the dense cellular environment. Existing sample preparation methods are insufficient at accessing the plasma membrane and its associated proteins. Here, we present a correlative cryo-electron tomography pipeline optimally suited to image large ultra-thin areas of isolated basal and apical plasma membranes. The pipeline allows for angstrom-scale structure determination with subtomogram averaging and employs a genetically encodable rapid chemically-induced electron microscopy visible tag for marking specific proteins within the complex cellular environment. The pipeline provides efficient, distributable, low-cost sample preparation and enables targeted structural studies of identified proteins at the plasma membrane of mammalian cells.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Update of
-
Cryo-electron tomography pipeline for plasma membranes.bioRxiv [Preprint]. 2024 Jun 28:2024.06.27.600657. doi: 10.1101/2024.06.27.600657. bioRxiv. 2024. Update in: Nat Commun. 2025 Jan 20;16(1):855. doi: 10.1038/s41467-025-56045-z. PMID: 39372776 Free PMC article. Updated. Preprint.
References
-
- Bäuerlein, F. J. & Baumeister, W. Towards visual proteomics at high resolution. J. Mol. Biol.433, 167187 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

