A single residue switch mediates the broad neutralization of Rotaviruses
- PMID: 39833145
- PMCID: PMC11746992
- DOI: 10.1038/s41467-025-56114-3
A single residue switch mediates the broad neutralization of Rotaviruses
Abstract
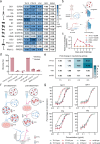
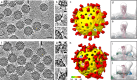
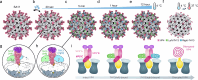
Broadly neutralizing antibodies (bNAbs) could offer escape-tolerant and lasting protection against viral infections and therefore guide development of broad-spectrum vaccines. The increasing challenge posed by viral evolution and immune evasion intensifies the importance of the discovery of bNAbs and their underlying neutralization mechanism. Here, focusing on the pivotal viral protein VP4 of rotavirus (RV), we identify a potent bNAb, 7H13, exhibiting broad-spectrum neutralization across diverse RV genotypes and demonstrating strong prevention of virus infection in female mice. A combination of time-resolved cryo-electron microscopy (cryo-EM) and in situ cryo-electron tomography (cryo-ET) analysis reveals a counterintuitive dynamic process of virus inactivation, in which 7H13 asymmetrically binds to a conserved epitope in the capsid-proximal aspect of VP4, triggers a conformational switch in a critical residue-F418-thereby disrupts the meta-stable conformation of VP4 essential for normal viral infection. Structure-guided mutagenesis corroborates the essential role of the 7H13 heavy chain I54 in activating F418 switch and destabilizing VP4. These findings define an atypical NAbs' neutralization mechanism and reveal a potential type of virus vulnerable site for universal vaccine and therapeutics design.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare that they have no conflicts of interest.
Figures


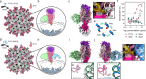
Similar articles
-
Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.J Virol. 2022 Aug 24;96(16):e0062722. doi: 10.1128/jvi.00627-22. Epub 2022 Aug 4. J Virol. 2022. PMID: 35924923 Free PMC article.
-
Expression and characterization of a novel truncated rotavirus VP4 for the development of a recombinant rotavirus vaccine.Vaccine. 2018 Apr 12;36(16):2086-2092. doi: 10.1016/j.vaccine.2018.03.011. Epub 2018 Mar 16. Vaccine. 2018. PMID: 29555220
-
Recombinant bivalent subunit vaccine combining truncated VP4 from P[7] and P[23] induces protective immunity against prevalent porcine rotaviruses.J Virol. 2024 May 14;98(5):e0021224. doi: 10.1128/jvi.00212-24. Epub 2024 Apr 9. J Virol. 2024. PMID: 38591886 Free PMC article.
-
Wa-VP4* as a candidate rotavirus vaccine induced homologous and heterologous virus neutralizing antibody responses in mice, pigs, and cynomolgus monkeys.Vaccine. 2024 May 31;42(15):3514-3521. doi: 10.1016/j.vaccine.2024.04.056. Epub 2024 Apr 25. Vaccine. 2024. PMID: 38670845
-
Reverse vaccinology-based multi-epitope vaccine design against Indian group A rotavirus targeting VP7, VP4, and VP6 proteins.Microb Pathog. 2024 Aug;193:106775. doi: 10.1016/j.micpath.2024.106775. Epub 2024 Jul 1. Microb Pathog. 2024. PMID: 38960216 Review.
Cited by
-
Engaging Broader Stakeholders to Accelerate Group A Streptococcus Vaccine Development.Vaccines (Basel). 2025 Jul 7;13(7):734. doi: 10.3390/vaccines13070734. Vaccines (Basel). 2025. PMID: 40733711 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

