Construction and iterative redesign of synXVI a 903 kb synthetic Saccharomyces cerevisiae chromosome
- PMID: 39833175
- PMCID: PMC11747415
- DOI: 10.1038/s41467-024-55318-3
Construction and iterative redesign of synXVI a 903 kb synthetic Saccharomyces cerevisiae chromosome
Abstract
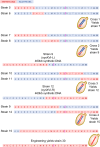
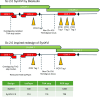
The Sc2.0 global consortium to design and construct a synthetic genome based on the Saccharomyces cerevisiae genome commenced in 2006, comprising 16 synthetic chromosomes and a new-to-nature tRNA neochromosome. In this paper we describe assembly and debugging of the 902,994-bp synthetic Saccharomyces cerevisiae chromosome synXVI of the Sc2.0 project. Application of the CRISPR D-BUGS protocol identified defective loci, which were modified to improve sporulation and recover wild-type like growth when grown on glycerol as a sole carbon source when grown at 37˚C. LoxPsym sites inserted downstream of dubious open reading frames impacted the 5' UTR of genes required for optimal growth and were identified as a systematic cause of defective growth. Based on lessons learned from analysis of Sc2.0 defects and synXVI, an in-silico redesign of the synXVI chromosome was performed, which can be used as a blueprint for future synthetic yeast genome designs. The in-silico redesign of synXVI includes reduced PCR tag frequency, modified chunk and megachunk termini, and adjustments to allocation of loxPsym sites and TAA stop codons to dubious ORFs. This redesign provides a roadmap into applications of Sc2.0 strategies in non-yeast organisms.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: T.C.W. and A.C.C. are founders and shareholders of Number 8 Bio Pty Ltd. J.D.B. is a Founder of and consultant to Opentrons LabWorks/Neochromosome, Inc, and serves or served on the Scientific Advisory Board of the following: CZ Biohub New York, LLC, Logomix, Inc., Modern Meadow, Inc., Rome Therapeutics, Inc., SeaHub, Seattle, WA, Tessera Therapeutics, Inc. and the Wyss Institute. J.S.B. is a Founder of Neochromosome, Inc., and a consultant to Opentrons Labworks, Inc. L.A.M. is a Founder of Neochromosome, Inc., and an employee of Opentrons Labworks, Inc. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

