Palmitoylation-dependent regulation of GPX4 suppresses ferroptosis
- PMID: 39833225
- PMCID: PMC11746948
- DOI: 10.1038/s41467-025-56344-5
Palmitoylation-dependent regulation of GPX4 suppresses ferroptosis
Abstract
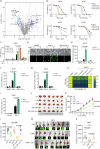
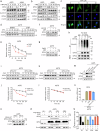
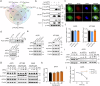
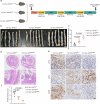
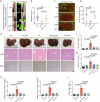
S-palmitoylation is a reversible and widespread post-translational modification, but its role in the regulation of ferroptosis has been poorly understood. Here, we elucidate that GPX4, an essential regulator of ferroptosis, is reversibly palmitoylated on cysteine 66. The acyltransferase ZDHHC20 palmitoylates GPX4 and increases its protein stability. ZDHHC20 depletion or inhibition of protein palmitoylation by 2-BP sensitizes cancer cells to ferroptosis. Moreover, we identify APT2 as the depalmitoylase of GPX4. Genetic silencing or pharmacological inhibition of APT2 with ML349 increases GPX4 palmitoylation, thereby stabilizing the protein and conferring resistance to ferroptosis. Notably, disrupting GPX4 palmitoylation markedly potentiates ferroptosis in xenografted and orthotopically implanted tumor models, and inhibits tumor metastasis through blood vessels. In the chemically induced colorectal cancer model, knockout of APT2 significantly aggravates cancer progression. Furthermore, pharmacologically modulating GPX4 palmitoylation impacts liver ischemia-reperfusion injury. Overall, our findings uncover the intricate network regulating GPX4 palmitoylation, highlighting its pivotal role in modulating ferroptosis sensitivity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

