Evolutionary analysis of the DHHCs in Saccharinae
- PMID: 39833334
- PMCID: PMC11756399
- DOI: 10.1038/s41598-025-86463-4
Evolutionary analysis of the DHHCs in Saccharinae
Abstract
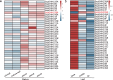
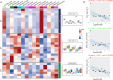
The DHHC domain genes are crucial for protein lipid modification, a key post-translational modification influencing membrane targeting, subcellular trafficking, and protein function. Despite their significance, the DHHC gene family in Saccharinae remains understudied. Here, we identified 32 (110 alleles), 28, 53, and 48 DHHC genes in Saccharum spontaneum Np-X, Erianthus rufipilus, Miscanthus sinensis, and Miscanthus lutarioriparius, respectively. Collinearity analysis uncovered the loss of two M. lutarioriparius genes, homologues of EruDHHC1C and EruDHHC3A. Phylogenetic and classification analyses categorized DHHC family members into six subgroups (A-F). Ka/Ks ratio analysis indicated that gene duplication in these species was primarily driven by whole-genome duplication (WGD) and dispersed duplication (DSD), with DHHC genes evolving under strong purifying selection. Gene expression and trait correlation analysis revealed a significant negative correlation between SspDHHC28A expression in S. spontaneum and sucrose content, suggesting a role in photosynthesis product transport during rapid growth. This study deepens our understanding of the DHHC gene family's functional dynamics and evolutionary path in Saccharinae, laying a foundation for future research.
Keywords: DHHC; Evolutionary; Gene duplications; Saccharinae; Sucrose storage.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Genome-wide systematic characterization of the HAK/KUP/KT gene family and its expression profile during plant growth and in response to low-K+ stress in Saccharum.BMC Plant Biol. 2020 Jan 13;20(1):20. doi: 10.1186/s12870-019-2227-7. BMC Plant Biol. 2020. PMID: 31931714 Free PMC article.
-
The reference genome of Miscanthus floridulus illuminates the evolution of Saccharinae.Nat Plants. 2021 May;7(5):608-618. doi: 10.1038/s41477-021-00908-y. Epub 2021 May 6. Nat Plants. 2021. PMID: 33958777 Free PMC article.
-
New insights into the evolution and functional divergence of the CIPK gene family in Saccharum.BMC Genomics. 2020 Dec 7;21(1):868. doi: 10.1186/s12864-020-07264-9. BMC Genomics. 2020. PMID: 33287700 Free PMC article.
-
Genome-Wide Identification and Characterization of CCT Gene Family from Microalgae to Legumes.Genes (Basel). 2024 Jul 18;15(7):941. doi: 10.3390/genes15070941. Genes (Basel). 2024. PMID: 39062720 Free PMC article.
-
Structure, phylogeny, allelic haplotypes and expression of sucrose transporter gene families in Saccharum.BMC Genomics. 2016 Feb 1;17:88. doi: 10.1186/s12864-016-2419-6. BMC Genomics. 2016. PMID: 26830680 Free PMC article.
References
-
- Greaves, J. & Chamberlain, L. H. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci.36, 245–253 (2011). - PubMed
-
- Putilina, T., Wong, P. & Gentleman, S. The DHHC domain: A new highly conserved cysteine-rich motif. - PubMed
-
- Mitchell, D. A., Vasudevan, A., Linder, M. E. & Deschenes, R. J. Thematic review series: lipid posttranslational modifications. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res.47, 1118–1127 (2006). - PubMed
-
- Hemsley, P. A. Protein S-acylation in plants (review). Mol. Membr. Biol.26, 114–125 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

