Systematic ocular phenotyping of 8,707 knockout mouse lines identifies genes associated with abnormal corneal phenotypes
- PMID: 39833678
- PMCID: PMC11744888
- DOI: 10.1186/s12864-025-11222-8
Systematic ocular phenotyping of 8,707 knockout mouse lines identifies genes associated with abnormal corneal phenotypes
Abstract
Purpose: Corneal dysmorphologies (CDs) are typically classified as either regressive degenerative corneal dystrophies (CDtrs) or defective growth and differentiation-driven corneal dysplasias (CDyps). Both eye disorders have multifactorial etiologies. While previous work has elucidated many aspects of CDs, such as presenting symptoms, epidemiology, and pathophysiology, the genetic mechanisms remain incompletely understood. The purpose of this study was to analyze phenotype data from 8,707 knockout mouse lines to identify new genes associated with the development of CDs in humans.
Methods: 8,707 knockout mouse lines phenotyped by the International Mouse Phenotyping Consortium were queried for genes associated with statistically significant (P < 0.0001) abnormal cornea morphology to identify candidate CD genes. Corneal abnormalities were investigated by histopathology. A literature search was used to determine the proportion of candidate genes previously associated with CDs in mice and humans. Phenotypes of human orthologues of mouse candidate genes were compared with known human CD genes to identify protein-protein interactions and molecular pathways using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), Protein Analysis Through Evolutionary Relationships (PANTHER), and Kyoto Encyclopedia of Genes and Genomes.
Results: Analysis of data from 8,707 knockout mouse lines identified 213 candidate CD genes. Of these, 37 (17%) genes were previously known to be associated with CD, including 14 in the mouse, 16 in humans, and 7 in both. The remaining 176 (83%) genes have not been previously implicated in CD. We also searched publicly available RNAseq data and found that 131 of the total 213 (61.5%) were expressed in adult human corneal tissue. STRING analysis showed several interactions within and between candidate and established CD proteins. All cellular pathways of the established genes were found in the PANTHER analysis of the candidate genes. Several of the candidate genes were implicated in corneal disease, such as TGF-ß signaling. We also identified other possible underappreciated mechanisms relevant to the human cornea.
Conclusions: We identified 213 mouse genes that resulted in statistically significant abnormal corneal phenotypes in knockout mice, many of which have not previously been implicated in corneal pathology. Bioinformatic analyses implicated candidate genes in several signaling pathways which are potential therapeutic targets.
Keywords: Corneal disease; Corneal dysmorphologies; Corneal dystrophies.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All procedures at each IMPC center adhered to local, state, and national regulatory guidelines, based on the standards of Animal Research: Reporting of In Vivo Experiments guidelines, a list of recommendations to standardize and improve the quality and reproducibility of animal research. A Housing and Husbandry protocol was also followed, which contains a collection of mandatory and optional procedures to be used during international mouse experimentation.4,5 Guidelines can be accessed at https://www.mousephenotype.org/about-impc/animal-welfare/ . All procedures on live animals were reviewed and approved by associated institutional animal care and use committees (IACUC), animal care committees (ACC), or equivalent. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
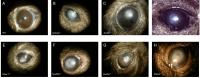



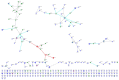


References
-
- Moshirfar M, Bennett P, Ronquillo Y. Corneal Dystrophy. In: StatPearls. StatPearls Publishing; 2024. Accessed April 17, 2024. http://www.ncbi.nlm.nih.gov/books/NBK557865/ - PubMed
-
- Elrick H, Peterson KA, Wood JA, et al. The production of 4,182 mouse lines identifies experimental and biological variables impacting Cas9-mediated mutant mouse line production. Published Online Oct. 2021;6. 2021.10.06.463037.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

