Fecal occult blood affects intestinal microbial community structure in colorectal cancer
- PMID: 39833681
- PMCID: PMC11745023
- DOI: 10.1186/s12866-024-03721-7
Fecal occult blood affects intestinal microbial community structure in colorectal cancer
Abstract
Background: Gut microbes have been used to predict CRC risk. Fecal occult blood test (FOBT) has been recommended for population screening of CRC.
Objective: To analyze the effects of fecal occult blood test (FOBT) on gut microbes.
Methods: Fecal samples from 107 healthy individuals (FOBT-negative) and 111 CRC patients (39 FOBT-negative and 72 FOBT-positive) were included for 16 S ribosomal RNA sequencing. Based on the results of different FOBT, the community structure and diversity of intestinal bacteria in healthy individuals and CRC patients were analyzed. Characteristic gut bacteria were screened, and various machine learning algorithms were applied to construct CRC risk prediction models.
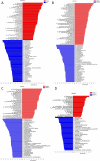
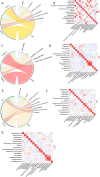
Results: The gut microbiota of healthy people and CRC patients with different fecal occult blood were mapped. There was no statistical difference in diversity between CRC patients with negative FOBT and positive FOBT. Bacteroides, Blautia and Escherichia-Shigella were more correlated to healthy individuals, while Streptococcus showed higher correlation with CRC patients with negative FOBT. The accuracy of CRC risk prediction model based on the support vector machines (SVM) algorithm was the highest (89.71%). Subsequently, FOBT was included as a characteristic element in the model construction, and the prediction accuracy of the model was all increased. Similarly, the CRC risk prediction model based on SVM algorithm had the highest accuracy (92%).
Conclusion: FOB affects the community composition of gut microbes. When predicting CRC risk based on gut microbiome, considering the influence of FOBT is expected to improve the accuracy of CRC risk prediction.
Keywords: 16S ribosomal RNA sequencing; Artificial intelligence; Colorectal cancer; Fecal occult blood test; Gut microbes; Risk prediction.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The clinical protocols involving the patients and the informed consent form were approved by the Chinese Clinical Trial Registry ( http://www.chictr.org.cn , ChiCTR1800018908) and Ethics Committee of Huzhou Central Hospital (No.202202005-01). All participants provided written informed consent. All methods were performed in accordance with the relevant guidelines and regulations in ethics approval and consent to participate. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

