Genetic context modulates aging and degeneration in the murine retina
- PMID: 39833899
- PMCID: PMC11744848
- DOI: 10.1186/s13024-025-00800-9
Genetic context modulates aging and degeneration in the murine retina
Abstract
Background: Age is the principal risk factor for neurodegeneration in both the retina and brain. The retina and brain share many biological properties; thus, insights into retinal aging and degeneration may shed light onto similar processes in the brain. Genetic makeup strongly influences susceptibility to age-related retinal disease. However, studies investigating retinal aging have not sufficiently accounted for genetic diversity. Therefore, examining molecular aging in the retina across different genetic backgrounds will enhance our understanding of human-relevant aging and degeneration in both the retina and brain-potentially improving therapeutic approaches to these debilitating conditions.
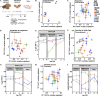
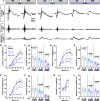
Methods: Transcriptomics and proteomics were employed to elucidate retinal aging signatures in nine genetically diverse mouse strains (C57BL/6J, 129S1/SvlmJ, NZO/HlLtJ, WSB/EiJ, CAST/EiJ, PWK/PhK, NOD/ShiLtJ, A/J, and BALB/cJ) across lifespan. These data predicted human disease-relevant changes in WSB and NZO strains. Accordingly, B6, WSB, and NZO mice were subjected to human-relevant in vivo examinations at 4, 8, 12, and/or 18M, including: slit lamp, fundus imaging, optical coherence tomography, fluorescein angiography, and pattern/full-field electroretinography. Retinal morphology, vascular structure, and cell counts were assessed ex vivo.
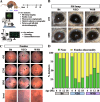
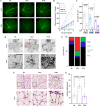
Results: We identified common molecular aging signatures across the nine mouse strains, which included genes associated with photoreceptor function and immune activation. Genetic background strongly modulated these aging signatures. Analysis of cell type-specific marker genes predicted age-related loss of photoreceptors and retinal ganglion cells (RGCs) in WSB and NZO, respectively. Fundus exams revealed retinitis pigmentosa-relevant pigmentary abnormalities in WSB retinas and diabetic retinopathy (DR)-relevant cotton wool spots and exudates in NZO retinas. Profound photoreceptor dysfunction and loss were confirmed in WSB. Molecular analyses indicated changes in photoreceptor-specific proteins prior to loss, suggesting photoreceptor-intrinsic dysfunction in WSB. In addition, age-associated RGC dysfunction, loss, and concomitant microvascular dysfunction were observed in NZO mice. Proteomic analyses revealed an early reduction in protective antioxidant processes, which may underlie increased susceptibility to DR-relevant pathology in NZO.
Conclusions: Genetic context is a strong determinant of retinal aging, and our multi-omics resource can aid in understanding age-related diseases of the eye and brain. Our investigations identified and validated WSB and NZO mice as improved preclinical models relevant to common retinal neurodegenerative diseases.
Keywords: Aging; Diabetic retinopathy; Genetic diversity; NZO; Proteomics; RNA-sequencing; Retina; Retinal vascular disease; Retinitis pigmentosa; WSB.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All research was approved by the Institutional Animal Care and Use Committee (IACUC) at The Jackson Laboratory. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures






Update of
-
Genetic context modulates aging and degeneration in the murine retina.bioRxiv [Preprint]. 2024 Apr 20:2024.04.16.589625. doi: 10.1101/2024.04.16.589625. bioRxiv. 2024. Update in: Mol Neurodegener. 2025 Jan 20;20(1):8. doi: 10.1186/s13024-025-00800-9. PMID: 38659747 Free PMC article. Updated. Preprint.
References
-
- Sabeti F, Lane J, Rohan EMF, Essex RW, McKone E, Maddess T. Relationships between retinal structure and function and vision-related quality of life measures in advanced age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2021;259(12):3687–96. - PubMed
-
- Wolfram C, Lorenz K, Breitscheidel L, Verboven Y, Pfeiffer N. Health- and vision-related quality of life in patients with ocular hypertension or primary open-angle glaucoma. Ophthalmologica. 2013;229(4):227–34. - PubMed
-
- Organization WH. World report on vision. 2019.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

