Differential regulation of apoptosis-related genes during long-term culture and differentiation of canine adipose-derived stem cells - a functional bioinformatical analysis
- PMID: 39834550
- PMCID: PMC11743971
- DOI: 10.3389/fgene.2024.1515778
Differential regulation of apoptosis-related genes during long-term culture and differentiation of canine adipose-derived stem cells - a functional bioinformatical analysis
Abstract
Introduction: Stem cells derived from adipose tissue are gaining popularity in the field of regenerative medicine due to their adaptability and clinical potential. Their rapid growth, ability to differentiate, and easy extraction with minimal complications make adipose-derived stem cells (ADSCs) a promising option for many treatments, particularly those targeting bone-related diseases. This study analyzed gene expression in canine ADSCs subjected to long-term culture and osteogenic differentiation.
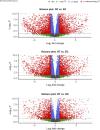
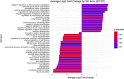
Methods: ADSCs were isolated from discarded surgical waste and cultured for 14 days with and without differentiation media to assess osteogenic changes. RNA sequencing (RNA-seq) and bioinformatical analysis were performed to obtain comprehensive transcriptomic data. A total of 17793 genes were detected and GO enrichment analysis was performed on the differentially expressed genes to identify significantly up- and downregulated Biological Process (BP) GO terms across each comparison.
Results: The upregulation of apoptosis-regulating genes and genes related to circulatory system development suggest an induction of these processes, while the downregulation of neurogenesis and gliogenesis genes points to reciprocal regulation during osteogenic differentiation of canine ADSCs.
Discussion: These findings underscore the potential of ADSCs in bone regeneration and offer valuable insights for advancing tissue engineering, however further studies, including proteomic analyses, are needed to confirm these patterns and their biological significance.
Keywords: adipose tissue; apoptosis; gene expression; gene ontology; stem cells; transcriptomics.
Copyright © 2025 Jankowski, Stefańska, Suchodolski, Dompe, Wąsiatycz, Kempisty, Nowicki and Roszak.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures












References
-
- Bieback K., Hecker A., Schlechter T., Hofmann I., Brousos N., Redmer T., et al. (2012). Replicative aging and differentiation potential of human adipose tissue-derived mesenchymal stromal cells expanded in pooled human or fetal bovine serum. Cytotherapy 14, 570–583. 10.3109/14653249.2011.652809 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

