LINC01305 and LAD1 Co-Regulate CTTN and N-WASP Phosphorylation, Mediating Cytoskeletal Reorganization to Promote ESCC Metastasis
- PMID: 39835575
- PMCID: PMC11890417
- DOI: 10.1002/mc.23885
LINC01305 and LAD1 Co-Regulate CTTN and N-WASP Phosphorylation, Mediating Cytoskeletal Reorganization to Promote ESCC Metastasis
Abstract
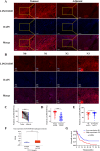
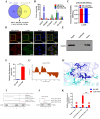
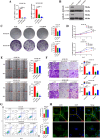
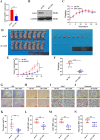
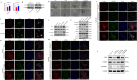
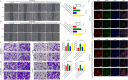
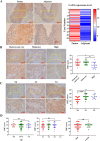
Esophageal squamous cell carcinoma (ESCC) is prone to metastasis and is a leading cause of mortality. The cytoskeleton is closely related to cell morphology and movement; however, little research has been conducted on ESCC metastasis. In this study, we found that the anchoring filament protein ladinin 1 (LAD1) specifically binds to LINC01305 for co-regulating the level of modulating cortactin proteins (CTTN) and neuronal Wiskott-Aldrich syndrome protein (N-WASP) phosphorylation, which mediates cytoskeletal reorganization and affects the metastasis of ESCC cells. Additionally, LINC01305 and LAD1 jointly promoted the epithelial-mesenchymal transition (EMT) process by activating the phosphoinositide-3-kinase-protein kinase B (PI3K/AKT) signaling pathway. Moreover, LINC01305 and LAD1 were related to the late clinical stage and lymph node metastasis of ESCC. Our study demonstrated that LINC01305 and LAD1 are major determinants of ESCC dissemination and revealed a novel molecular mechanism of cytoskeletal reorganization that controls ESCC metastasis. Trial Registration: N/A.
Keywords: ESCC; LAD1; LINC01305; cytoskeleton; metastasis.
© 2025 The Author(s). Molecular Carcinogenesis published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
linc01305 promotes metastasis and proliferation of esophageal squamous cell carcinoma through interacting with IGF2BP2 and IGF2BP3 to stabilize HTR3A mRNA.Int J Biochem Cell Biol. 2021 Jul;136:106015. doi: 10.1016/j.biocel.2021.106015. Epub 2021 May 19. Int J Biochem Cell Biol. 2021. PMID: 34022433
-
Long non‑coding RNA MEG3 suppresses epithelial‑to‑mesenchymal transition by inhibiting the PSAT1‑dependent GSK‑3β/Snail signaling pathway in esophageal squamous cell carcinoma.Oncol Rep. 2020 Nov;44(5):2130-2142. doi: 10.3892/or.2020.7754. Epub 2020 Sep 7. Oncol Rep. 2020. PMID: 32901893 Free PMC article.
-
Long noncoding RNA SNHG12 induces proliferation, migration, epithelial-mesenchymal transition, and stemness of esophageal squamous cell carcinoma cells via post-transcriptional regulation of BMI1 and CTNNB1.Mol Oncol. 2020 Sep;14(9):2332-2351. doi: 10.1002/1878-0261.12683. Epub 2020 Jun 18. Mol Oncol. 2020. PMID: 32239639 Free PMC article.
-
A novel long noncoding RNA, LOC440173, promotes the progression of esophageal squamous cell carcinoma by modulating the miR-30d-5p/HDAC9 axis and the epithelial-mesenchymal transition.Mol Carcinog. 2020 Dec;59(12):1392-1408. doi: 10.1002/mc.23264. Epub 2020 Oct 20. Mol Carcinog. 2020. PMID: 33079409
-
Roles of long non‑coding RNAs in esophageal cell squamous carcinoma (Review).Int J Mol Med. 2024 Aug;54(2):72. doi: 10.3892/ijmm.2024.5396. Epub 2024 Jul 4. Int J Mol Med. 2024. PMID: 38963019 Free PMC article. Review.
References
-
- Zhang Y. and Chen Y., “Stratification From Heterogeneity of the Cell‐Death Signal Enables Prognosis Prediction and Immune Microenvironment Characterization in Esophageal Squamous Cell Carcinoma,” Frontiers in Cell and Developmental Biology 10 (2022): 855404, 10.3389/fcell.2022.855404. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

