RNASEK interacting with PEDV structural proteins facilitates virus entry via clathrin-mediated endocytosis
- PMID: 39835814
- PMCID: PMC11852855
- DOI: 10.1128/jvi.01760-24
RNASEK interacting with PEDV structural proteins facilitates virus entry via clathrin-mediated endocytosis
Abstract
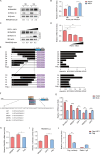
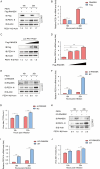
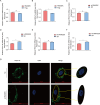
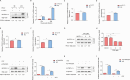
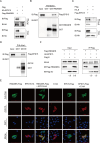
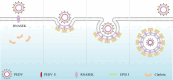
Porcine epidemic diarrhea virus (PEDV), as a type of Alphacoronavirus causing acute diarrhea and high death rate among sucking piglets, poses great financial damage to the swine industry. Nevertheless, the molecular mechanism whereby PEDV enters host cells is unclear, limiting the development of PED vaccines and anti-PEDV agents. The present study found that the host protein ribonuclease kappa (RNASEK) was regulated by USF2, a transcription factor, and facilitated the PEDV replication. RNASEK was identified as a novel binding partner of PEDV, which interacted with a spike (S), envelope (E), and membrane (M) proteins on PEDV virion surfaces to increase the uptake not for attachment of PEDV virions. PEDV enters cells through the endocytosis pathways. RNASEK knockdown or RNASEK knockout assay revealed that through clathrin-mediated endocytosis (CME), RNASEK promoted the internalization of PEDV virions. Clathrin and the adaptor protein EPS15 only interacted with PEDV E protein, demonstrating that the RNASEK could target more virions through interaction with PEDV S, E, and M proteins to clathrin and EPS15 proteins rather than merely interacting with PEDV E protein to mediate the PEDV entry through CME. Moreover, our findings suggest that RNASEK, a newly identified host-entry factor, facilitates PEDV internalization by increasing the interaction of PEDV virions and EPS15-clathrin complex and may also provide a potential target for anti-PEDV therapies.IMPORTANCEPEDV is the causative pathogen of porcine diarrhea, which is a highly infectious acute intestinal condition, that poses significant economic damage to the swine industry. However, the existing PED vaccines fail to provide adequate protection for piglets against PEDV infection. Although PEDV replication in cells has been widely described, the mechanisms beneath PEDV entry of the host cells are incompletely understood. In this study, we showed that RNASEK, regulated by the transcription factor USF2, is a new host factor increasing PEDV infection in LLC-PK1 cells. RNASEK can bind to multiple structural proteins of PEDV (S, E, and M proteins), therefore increasing the interaction between PEDV virions, clathrin, and EPS15 to promote PEDV virion entry. Apart from unraveling the entry mechanisms of PEDV, our findings also contributed to facilitating the development of anti-PEDV agents and PED vaccines.
Keywords: PEDV; RNASEK; endocytosis; virus entry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Clathrin- and serine proteases-dependent uptake of porcine epidemic diarrhea virus into Vero cells.Virus Res. 2014 Oct 13;191:21-9. doi: 10.1016/j.virusres.2014.07.022. Epub 2014 Jul 31. Virus Res. 2014. PMID: 25086180 Free PMC article.
-
Dynamic Dissection of the Endocytosis of Porcine Epidemic Diarrhea Coronavirus Cooperatively Mediated by Clathrin and Caveolae as Visualized by Single-Virus Tracking.mBio. 2021 Mar 30;12(2):e00256-21. doi: 10.1128/mBio.00256-21. mBio. 2021. PMID: 33785615 Free PMC article.
-
FSTL1 and TLR4 interact with PEDV structural proteins to promote virus adsorption to host cells.J Virol. 2025 Jan 31;99(1):e0183724. doi: 10.1128/jvi.01837-24. Epub 2024 Dec 13. J Virol. 2025. PMID: 39670742 Free PMC article.
-
Cellular entry of the porcine epidemic diarrhea virus.Virus Res. 2016 Dec 2;226:117-127. doi: 10.1016/j.virusres.2016.05.031. Epub 2016 Jun 15. Virus Res. 2016. PMID: 27317167 Free PMC article. Review.
-
The biomechanical phenomena observed in the cell invasion pathway of porcine epidemic diarrhea virus: a review.Arch Virol. 2025 May 26;170(7):139. doi: 10.1007/s00705-025-06326-1. Arch Virol. 2025. PMID: 40418401 Review.
References
MeSH terms
Substances
Grants and funding
- 2023YFF1000900/MOST | National Key Research and Development Program of China (NKPs)
- 2023YFD1801300/MOST | National Key Research and Development Program of China (NKPs)
- 32272999/MOST | National Natural Science Foundation of China (NSFC)
- 23ZR1476900/STCSM | Natural Science Foundation of Shanghai Municipality ()
LinkOut - more resources
Full Text Sources
Miscellaneous

